
Culex quinquefasciatus (Southern house mosquito) (Culex pungens)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Nematocera;
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
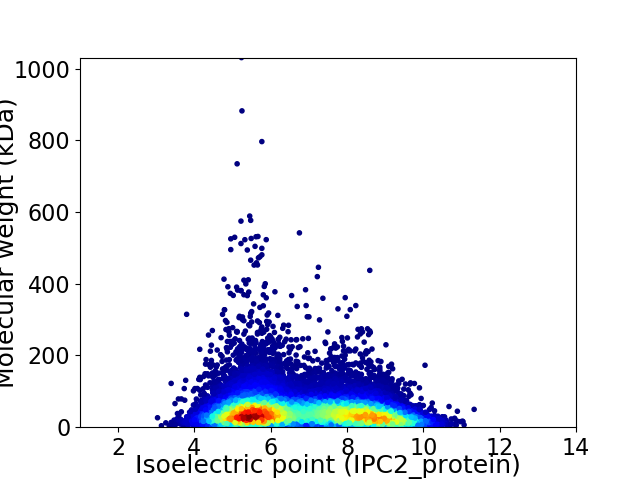
Virtual 2D-PAGE plot for 19204 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
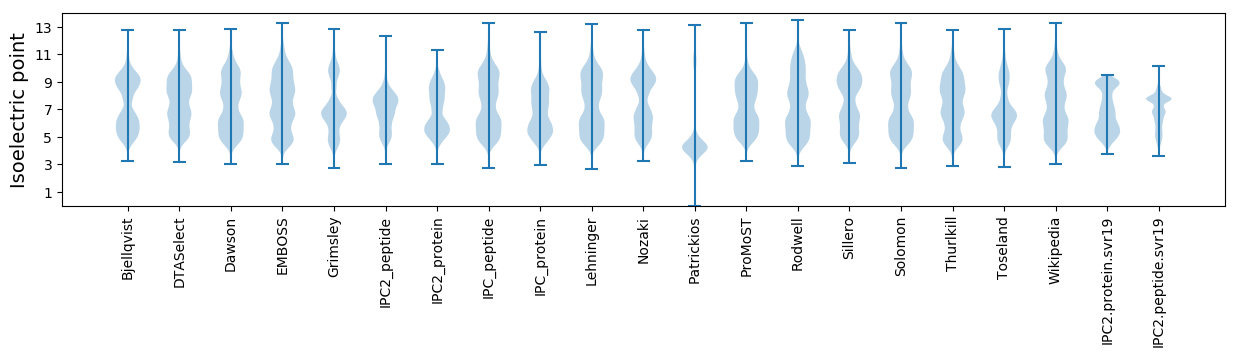
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B0X7D9|B0X7D9_CULQU Tryptophanyl-tRNA synthetase OS=Culex quinquefasciatus OX=7176 GN=6048677 PE=3 SV=1
MM1 pKa = 7.35SLCKK5 pKa = 10.52AFGKK9 pKa = 10.49DD10 pKa = 3.01KK11 pKa = 11.29VIFATAEE18 pKa = 3.82DD19 pKa = 4.29HH20 pKa = 6.25ATPSTVQLPEE30 pKa = 4.58SEE32 pKa = 4.34PQPGLILDD40 pKa = 4.29NGDD43 pKa = 3.8INWNCPCLGGMATGPCGVEE62 pKa = 3.76FRR64 pKa = 11.84EE65 pKa = 4.39AFSCFHH71 pKa = 6.44YY72 pKa = 10.96SEE74 pKa = 4.93AAPKK78 pKa = 10.83GSDD81 pKa = 3.62CYY83 pKa = 11.37DD84 pKa = 3.28AFKK87 pKa = 10.77TMQDD91 pKa = 3.08CMGNYY96 pKa = 8.81PGVYY100 pKa = 9.77AKK102 pKa = 10.45QNQRR106 pKa = 11.84DD107 pKa = 3.79GDD109 pKa = 4.32DD110 pKa = 4.3DD111 pKa = 6.35DD112 pKa = 6.23EE113 pKa = 7.95DD114 pKa = 5.91DD115 pKa = 5.44GLDD118 pKa = 4.29LGAAMAGAEE127 pKa = 4.18QEE129 pKa = 4.76SPDD132 pKa = 3.96KK133 pKa = 11.44VSVDD137 pKa = 3.7SVDD140 pKa = 3.33ATDD143 pKa = 4.4AGSSAAVVQKK153 pKa = 10.9SVV155 pKa = 2.73
MM1 pKa = 7.35SLCKK5 pKa = 10.52AFGKK9 pKa = 10.49DD10 pKa = 3.01KK11 pKa = 11.29VIFATAEE18 pKa = 3.82DD19 pKa = 4.29HH20 pKa = 6.25ATPSTVQLPEE30 pKa = 4.58SEE32 pKa = 4.34PQPGLILDD40 pKa = 4.29NGDD43 pKa = 3.8INWNCPCLGGMATGPCGVEE62 pKa = 3.76FRR64 pKa = 11.84EE65 pKa = 4.39AFSCFHH71 pKa = 6.44YY72 pKa = 10.96SEE74 pKa = 4.93AAPKK78 pKa = 10.83GSDD81 pKa = 3.62CYY83 pKa = 11.37DD84 pKa = 3.28AFKK87 pKa = 10.77TMQDD91 pKa = 3.08CMGNYY96 pKa = 8.81PGVYY100 pKa = 9.77AKK102 pKa = 10.45QNQRR106 pKa = 11.84DD107 pKa = 3.79GDD109 pKa = 4.32DD110 pKa = 4.3DD111 pKa = 6.35DD112 pKa = 6.23EE113 pKa = 7.95DD114 pKa = 5.91DD115 pKa = 5.44GLDD118 pKa = 4.29LGAAMAGAEE127 pKa = 4.18QEE129 pKa = 4.76SPDD132 pKa = 3.96KK133 pKa = 11.44VSVDD137 pKa = 3.7SVDD140 pKa = 3.33ATDD143 pKa = 4.4AGSSAAVVQKK153 pKa = 10.9SVV155 pKa = 2.73
Molecular weight: 16.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B0WKZ1|B0WKZ1_CULQU DUF4774 domain-containing protein OS=Culex quinquefasciatus OX=7176 GN=6039874 PE=4 SV=1
MM1 pKa = 7.37TMAKK5 pKa = 10.2LATLLCWLRR14 pKa = 11.84SMNLPPNRR22 pKa = 11.84NRR24 pKa = 11.84PASPSQNQRR33 pKa = 11.84VNPSRR38 pKa = 11.84LVNRR42 pKa = 11.84NPLVNRR48 pKa = 11.84SLLVNRR54 pKa = 11.84NPLASLHH61 pKa = 6.08LQVNPNQWANRR72 pKa = 11.84SQQVNLNQRR81 pKa = 11.84ASQNQQVNLSPRR93 pKa = 11.84VNQKK97 pKa = 10.77LLVNPNQLASLNPTVNPSRR116 pKa = 11.84PVNLNRR122 pKa = 11.84RR123 pKa = 11.84VNSSQQVNLRR133 pKa = 11.84LPLSS137 pKa = 3.73
MM1 pKa = 7.37TMAKK5 pKa = 10.2LATLLCWLRR14 pKa = 11.84SMNLPPNRR22 pKa = 11.84NRR24 pKa = 11.84PASPSQNQRR33 pKa = 11.84VNPSRR38 pKa = 11.84LVNRR42 pKa = 11.84NPLVNRR48 pKa = 11.84SLLVNRR54 pKa = 11.84NPLASLHH61 pKa = 6.08LQVNPNQWANRR72 pKa = 11.84SQQVNLNQRR81 pKa = 11.84ASQNQQVNLSPRR93 pKa = 11.84VNQKK97 pKa = 10.77LLVNPNQLASLNPTVNPSRR116 pKa = 11.84PVNLNRR122 pKa = 11.84RR123 pKa = 11.84VNSSQQVNLRR133 pKa = 11.84LPLSS137 pKa = 3.73
Molecular weight: 15.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8459113 |
29 |
9108 |
440.5 |
49.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.989 ± 0.019 | 2.037 ± 0.018 |
5.345 ± 0.013 | 6.48 ± 0.023 |
3.948 ± 0.016 | 6.155 ± 0.021 |
2.525 ± 0.011 | 5.059 ± 0.014 |
5.865 ± 0.023 | 8.973 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.292 ± 0.009 | 4.604 ± 0.012 |
5.324 ± 0.024 | 4.544 ± 0.022 |
5.685 ± 0.015 | 7.924 ± 0.025 |
5.722 ± 0.015 | 6.471 ± 0.016 |
1.059 ± 0.007 | 2.998 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
