
Pseudogymnoascus sp. VKM F-3557
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Leotiomycetes incertae sedis; Pseudeurotiaceae; Pseudogymnoascus; unclassified Pseudogymnoascus
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
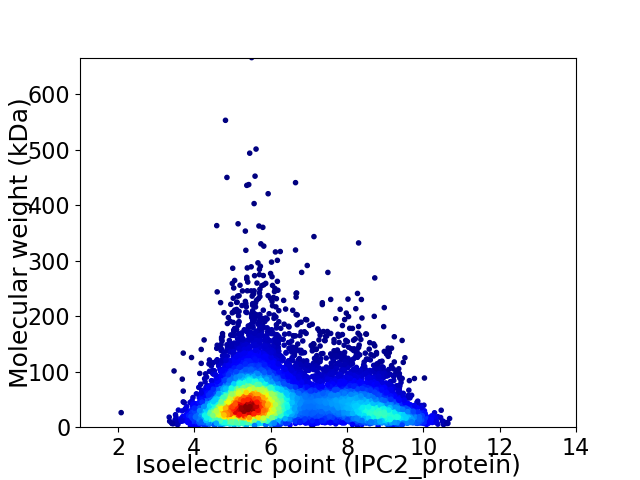
Virtual 2D-PAGE plot for 9480 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A093XV64|A0A093XV64_9PEZI Uncharacterized protein OS=Pseudogymnoascus sp. VKM F-3557 OX=1437433 GN=V490_03263 PE=3 SV=1
MM1 pKa = 7.25VFVKK5 pKa = 10.47PIFALSLVAGLIAPALSAAVRR26 pKa = 11.84INALGDD32 pKa = 4.11SITGSPGCWRR42 pKa = 11.84ALLYY46 pKa = 10.58QKK48 pKa = 10.63LVEE51 pKa = 4.74ANITDD56 pKa = 3.34IDD58 pKa = 4.23FVGTLPGQGCGIEE71 pKa = 3.98YY72 pKa = 10.6DD73 pKa = 4.23GEE75 pKa = 4.34NDD77 pKa = 3.27GHH79 pKa = 7.39GGFLATGIVADD90 pKa = 3.97NQLPGWLAVSQPDD103 pKa = 3.76VVMMQLATNDD113 pKa = 3.04VWSNIATATILDD125 pKa = 4.08AFSTLVDD132 pKa = 3.18QMRR135 pKa = 11.84DD136 pKa = 3.31SKK138 pKa = 10.74STMHH142 pKa = 6.54IVVAQITPMNPTDD155 pKa = 3.49GCATCEE161 pKa = 3.88AGIIALNDD169 pKa = 5.16AIPAWAEE176 pKa = 3.96EE177 pKa = 4.27KK178 pKa = 9.85STTEE182 pKa = 3.79SPITVVDD189 pKa = 4.99CYY191 pKa = 10.57TGYY194 pKa = 8.59DD195 pKa = 3.85TATDD199 pKa = 3.96TYY201 pKa = 11.53DD202 pKa = 3.5GVHH205 pKa = 6.75PNDD208 pKa = 3.7SGNAKK213 pKa = 9.66LAAAWFEE220 pKa = 4.13PLSAAIAAASSS231 pKa = 3.29
MM1 pKa = 7.25VFVKK5 pKa = 10.47PIFALSLVAGLIAPALSAAVRR26 pKa = 11.84INALGDD32 pKa = 4.11SITGSPGCWRR42 pKa = 11.84ALLYY46 pKa = 10.58QKK48 pKa = 10.63LVEE51 pKa = 4.74ANITDD56 pKa = 3.34IDD58 pKa = 4.23FVGTLPGQGCGIEE71 pKa = 3.98YY72 pKa = 10.6DD73 pKa = 4.23GEE75 pKa = 4.34NDD77 pKa = 3.27GHH79 pKa = 7.39GGFLATGIVADD90 pKa = 3.97NQLPGWLAVSQPDD103 pKa = 3.76VVMMQLATNDD113 pKa = 3.04VWSNIATATILDD125 pKa = 4.08AFSTLVDD132 pKa = 3.18QMRR135 pKa = 11.84DD136 pKa = 3.31SKK138 pKa = 10.74STMHH142 pKa = 6.54IVVAQITPMNPTDD155 pKa = 3.49GCATCEE161 pKa = 3.88AGIIALNDD169 pKa = 5.16AIPAWAEE176 pKa = 3.96EE177 pKa = 4.27KK178 pKa = 9.85STTEE182 pKa = 3.79SPITVVDD189 pKa = 4.99CYY191 pKa = 10.57TGYY194 pKa = 8.59DD195 pKa = 3.85TATDD199 pKa = 3.96TYY201 pKa = 11.53DD202 pKa = 3.5GVHH205 pKa = 6.75PNDD208 pKa = 3.7SGNAKK213 pKa = 9.66LAAAWFEE220 pKa = 4.13PLSAAIAAASSS231 pKa = 3.29
Molecular weight: 24.08 kDa
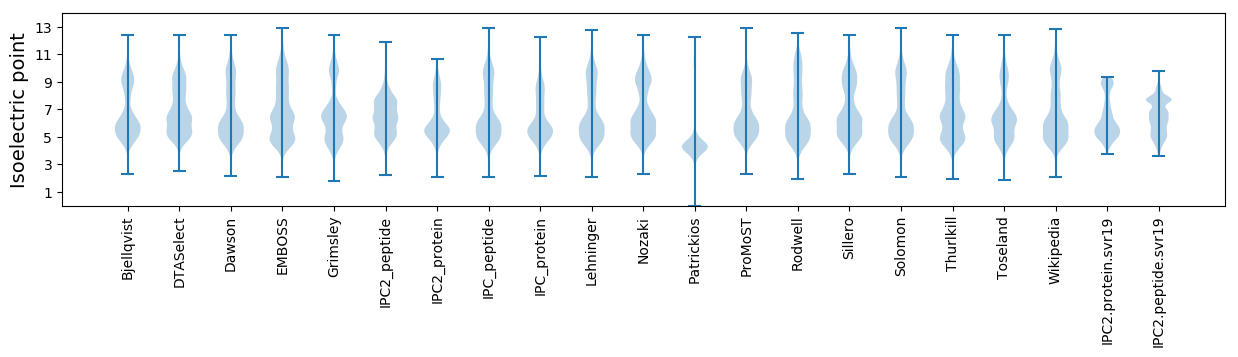
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A093XCJ2|A0A093XCJ2_9PEZI Uncharacterized protein OS=Pseudogymnoascus sp. VKM F-3557 OX=1437433 GN=V490_06485 PE=4 SV=1
MM1 pKa = 7.13GHH3 pKa = 6.44SNQGPAGDD11 pKa = 4.45GGSGGAICGGVGRR24 pKa = 11.84LAMQGHH30 pKa = 7.92LLPVDD35 pKa = 4.51SIGGHH40 pKa = 6.26HH41 pKa = 7.29PPPGPYY47 pKa = 9.97FGTDD51 pKa = 3.16PQIFPRR57 pKa = 11.84TAVEE61 pKa = 4.19DD62 pKa = 3.99ADD64 pKa = 3.55AMRR67 pKa = 11.84YY68 pKa = 9.83RR69 pKa = 11.84MLALQAPAVPKK80 pKa = 10.84APAARR85 pKa = 11.84NAPAAQWQNAMPAPLQTSGQSWQTAPAQSMQGHH118 pKa = 5.51MHH120 pKa = 6.69NYY122 pKa = 9.96DD123 pKa = 3.56HH124 pKa = 7.29TSPTPPPKK132 pKa = 9.66RR133 pKa = 11.84SRR135 pKa = 11.84PSRR138 pKa = 11.84PRR140 pKa = 11.84QQPRR144 pKa = 11.84QQAHH148 pKa = 4.81QRR150 pKa = 11.84AATHH154 pKa = 5.64PQIHH158 pKa = 5.57TTIQHH163 pKa = 6.14APVLQAPAQQPGLQASAQQVGPHH186 pKa = 5.82FVQLSTCNCLRR197 pKa = 11.84PISQLSLATPTSFDD211 pKa = 3.19SDD213 pKa = 3.68FFRR216 pKa = 11.84LSACACNILQLSVTFCLRR234 pKa = 11.84LLTMSRR240 pKa = 11.84SFSRR244 pKa = 11.84LPAASLHH251 pKa = 5.81LQTSRR256 pKa = 11.84VQPVLITFILQKK268 pKa = 10.4KK269 pKa = 8.48IKK271 pKa = 9.76NN272 pKa = 3.62
MM1 pKa = 7.13GHH3 pKa = 6.44SNQGPAGDD11 pKa = 4.45GGSGGAICGGVGRR24 pKa = 11.84LAMQGHH30 pKa = 7.92LLPVDD35 pKa = 4.51SIGGHH40 pKa = 6.26HH41 pKa = 7.29PPPGPYY47 pKa = 9.97FGTDD51 pKa = 3.16PQIFPRR57 pKa = 11.84TAVEE61 pKa = 4.19DD62 pKa = 3.99ADD64 pKa = 3.55AMRR67 pKa = 11.84YY68 pKa = 9.83RR69 pKa = 11.84MLALQAPAVPKK80 pKa = 10.84APAARR85 pKa = 11.84NAPAAQWQNAMPAPLQTSGQSWQTAPAQSMQGHH118 pKa = 5.51MHH120 pKa = 6.69NYY122 pKa = 9.96DD123 pKa = 3.56HH124 pKa = 7.29TSPTPPPKK132 pKa = 9.66RR133 pKa = 11.84SRR135 pKa = 11.84PSRR138 pKa = 11.84PRR140 pKa = 11.84QQPRR144 pKa = 11.84QQAHH148 pKa = 4.81QRR150 pKa = 11.84AATHH154 pKa = 5.64PQIHH158 pKa = 5.57TTIQHH163 pKa = 6.14APVLQAPAQQPGLQASAQQVGPHH186 pKa = 5.82FVQLSTCNCLRR197 pKa = 11.84PISQLSLATPTSFDD211 pKa = 3.19SDD213 pKa = 3.68FFRR216 pKa = 11.84LSACACNILQLSVTFCLRR234 pKa = 11.84LLTMSRR240 pKa = 11.84SFSRR244 pKa = 11.84LPAASLHH251 pKa = 5.81LQTSRR256 pKa = 11.84VQPVLITFILQKK268 pKa = 10.4KK269 pKa = 8.48IKK271 pKa = 9.76NN272 pKa = 3.62
Molecular weight: 29.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4561027 |
8 |
5968 |
481.1 |
53.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.88 ± 0.027 | 1.209 ± 0.011 |
5.674 ± 0.023 | 6.308 ± 0.027 |
3.649 ± 0.014 | 7.17 ± 0.023 |
2.233 ± 0.01 | 5.112 ± 0.017 |
5.095 ± 0.022 | 8.74 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.158 ± 0.009 | 3.78 ± 0.014 |
5.856 ± 0.024 | 3.865 ± 0.022 |
5.769 ± 0.023 | 8.112 ± 0.027 |
6.05 ± 0.019 | 6.138 ± 0.019 |
1.415 ± 0.009 | 2.788 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
