
Streptococcus satellite phage Javan260
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
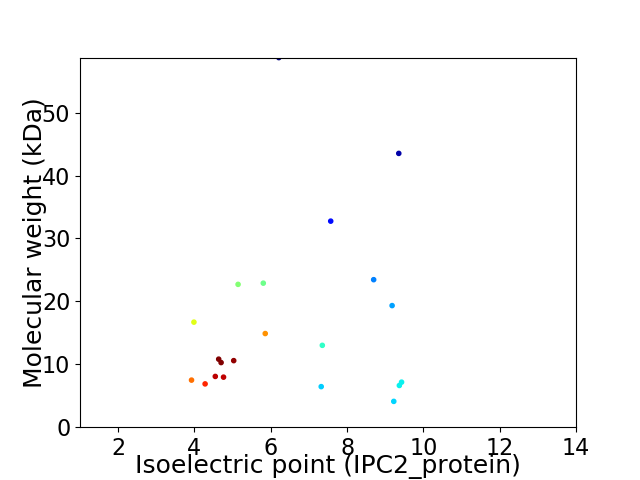
Virtual 2D-PAGE plot for 21 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZJI8|A0A4D5ZJI8_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan260 OX=2558596 GN=JavanS260_0011 PE=4 SV=1
MM1 pKa = 7.39LVTYY5 pKa = 8.27PALFYY10 pKa = 11.35YY11 pKa = 10.68DD12 pKa = 4.04DD13 pKa = 4.01TDD15 pKa = 3.86GANAPYY21 pKa = 10.03FVTFPDD27 pKa = 4.61FEE29 pKa = 5.3HH30 pKa = 7.07SATQGEE36 pKa = 4.54DD37 pKa = 3.02MANAMAMASDD47 pKa = 3.31WLGIHH52 pKa = 6.56LADD55 pKa = 4.36YY56 pKa = 10.61IEE58 pKa = 4.46NGRR61 pKa = 11.84DD62 pKa = 3.13IPTPAPINTLSLADD76 pKa = 3.94NNPFRR81 pKa = 11.84DD82 pKa = 3.99DD83 pKa = 3.55EE84 pKa = 5.07DD85 pKa = 4.4IEE87 pKa = 5.57LIYY90 pKa = 10.9DD91 pKa = 3.72PSKK94 pKa = 11.33SFISMVMVDD103 pKa = 3.25VAEE106 pKa = 4.14YY107 pKa = 10.49LGSQEE112 pKa = 4.15PVKK115 pKa = 10.18KK116 pKa = 9.12TLTIPRR122 pKa = 11.84WADD125 pKa = 3.11TLGRR129 pKa = 11.84DD130 pKa = 3.39LGLNFSQTLTDD141 pKa = 5.45AIADD145 pKa = 4.03KK146 pKa = 10.56KK147 pKa = 10.31IHH149 pKa = 5.92AA150 pKa = 5.0
MM1 pKa = 7.39LVTYY5 pKa = 8.27PALFYY10 pKa = 11.35YY11 pKa = 10.68DD12 pKa = 4.04DD13 pKa = 4.01TDD15 pKa = 3.86GANAPYY21 pKa = 10.03FVTFPDD27 pKa = 4.61FEE29 pKa = 5.3HH30 pKa = 7.07SATQGEE36 pKa = 4.54DD37 pKa = 3.02MANAMAMASDD47 pKa = 3.31WLGIHH52 pKa = 6.56LADD55 pKa = 4.36YY56 pKa = 10.61IEE58 pKa = 4.46NGRR61 pKa = 11.84DD62 pKa = 3.13IPTPAPINTLSLADD76 pKa = 3.94NNPFRR81 pKa = 11.84DD82 pKa = 3.99DD83 pKa = 3.55EE84 pKa = 5.07DD85 pKa = 4.4IEE87 pKa = 5.57LIYY90 pKa = 10.9DD91 pKa = 3.72PSKK94 pKa = 11.33SFISMVMVDD103 pKa = 3.25VAEE106 pKa = 4.14YY107 pKa = 10.49LGSQEE112 pKa = 4.15PVKK115 pKa = 10.18KK116 pKa = 9.12TLTIPRR122 pKa = 11.84WADD125 pKa = 3.11TLGRR129 pKa = 11.84DD130 pKa = 3.39LGLNFSQTLTDD141 pKa = 5.45AIADD145 pKa = 4.03KK146 pKa = 10.56KK147 pKa = 10.31IHH149 pKa = 5.92AA150 pKa = 5.0
Molecular weight: 16.7 kDa
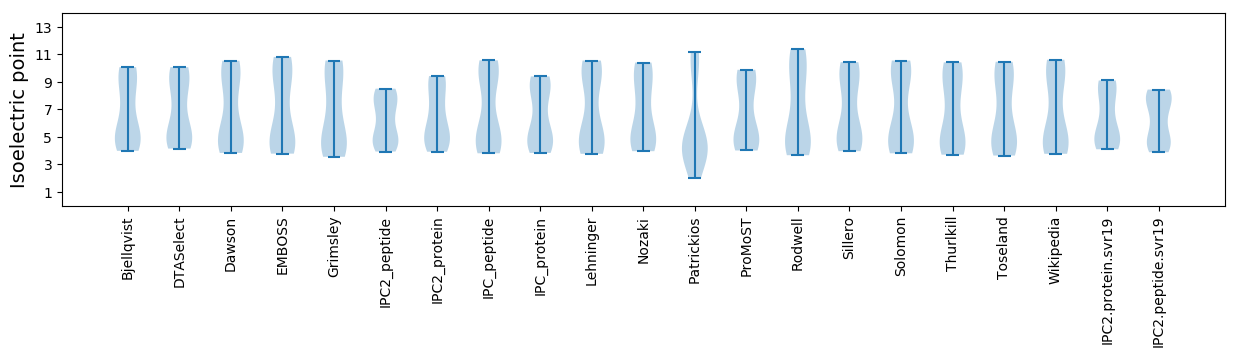
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZM32|A0A4D5ZM32_9VIRU DNA primase OS=Streptococcus satellite phage Javan260 OX=2558596 GN=JavanS260_0006 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 10.09IKK4 pKa = 10.66PITKK8 pKa = 9.17KK9 pKa = 10.51NGAIVYY15 pKa = 8.3RR16 pKa = 11.84ANVYY20 pKa = 10.65LGTDD24 pKa = 2.95QVTGKK29 pKa = 9.97KK30 pKa = 10.33AKK32 pKa = 9.51TSVTGRR38 pKa = 11.84TRR40 pKa = 11.84KK41 pKa = 8.76EE42 pKa = 3.84VKK44 pKa = 10.02QKK46 pKa = 10.96AKK48 pKa = 10.04HH49 pKa = 5.68AQDD52 pKa = 3.62EE53 pKa = 5.21FISNGYY59 pKa = 7.35TVTKK63 pKa = 9.88VVPIKK68 pKa = 10.67NYY70 pKa = 10.29QEE72 pKa = 4.68LAEE75 pKa = 4.74LWLEE79 pKa = 4.32SYY81 pKa = 10.85QLTVKK86 pKa = 9.85PQTFIATKK94 pKa = 10.24RR95 pKa = 11.84MLYY98 pKa = 10.01NHH100 pKa = 7.89LIPVFGALKK109 pKa = 10.32LNKK112 pKa = 10.01LSVSYY117 pKa = 10.64IQGFINDD124 pKa = 4.13LSAEE128 pKa = 4.15LVHH131 pKa = 6.38YY132 pKa = 10.48SVVHH136 pKa = 5.68SINRR140 pKa = 11.84RR141 pKa = 11.84VLQYY145 pKa = 10.88GVSLQLLPFNPARR158 pKa = 11.84DD159 pKa = 3.54VMLPKK164 pKa = 10.25VPKK167 pKa = 10.2KK168 pKa = 9.64EE169 pKa = 3.73NKK171 pKa = 9.48AIKK174 pKa = 10.16FIAPEE179 pKa = 3.97DD180 pKa = 3.88LKK182 pKa = 11.69ALMAYY187 pKa = 8.24MEE189 pKa = 4.47KK190 pKa = 10.52LANKK194 pKa = 9.07KK195 pKa = 9.8FSYY198 pKa = 10.51FFDD201 pKa = 3.67YY202 pKa = 11.08VLYY205 pKa = 10.68SVLLATGCRR214 pKa = 11.84FGEE217 pKa = 4.58VVALEE222 pKa = 4.12WSDD225 pKa = 3.32IDD227 pKa = 5.09LEE229 pKa = 4.52NGTISITKK237 pKa = 9.93NYY239 pKa = 10.09SRR241 pKa = 11.84LLKK244 pKa = 10.89LIGTPKK250 pKa = 9.91SKK252 pKa = 10.87AGVRR256 pKa = 11.84VISIDD261 pKa = 3.23KK262 pKa = 9.23KK263 pKa = 9.18TINLLRR269 pKa = 11.84LYY271 pKa = 10.58KK272 pKa = 10.31NRR274 pKa = 11.84QRR276 pKa = 11.84QIFIEE281 pKa = 4.08TGARR285 pKa = 11.84VSAVVFATPTKK296 pKa = 9.74EE297 pKa = 4.11YY298 pKa = 10.79QNMATRR304 pKa = 11.84QEE306 pKa = 4.21SLDD309 pKa = 3.62RR310 pKa = 11.84RR311 pKa = 11.84ITEE314 pKa = 3.41IGIPRR319 pKa = 11.84FTFHH323 pKa = 7.74AFRR326 pKa = 11.84HH327 pKa = 4.69THH329 pKa = 6.92ASLLLNAGISYY340 pKa = 10.68KK341 pKa = 10.23EE342 pKa = 3.61LQYY345 pKa = 11.49RR346 pKa = 11.84LGHH349 pKa = 6.19ATLAMTMDD357 pKa = 4.28IYY359 pKa = 11.69SHH361 pKa = 7.7LSMDD365 pKa = 3.69KK366 pKa = 10.11EE367 pKa = 4.32KK368 pKa = 10.95EE369 pKa = 3.93AVSYY373 pKa = 9.5YY374 pKa = 10.62EE375 pKa = 4.12KK376 pKa = 10.7AINSLL381 pKa = 3.57
MM1 pKa = 7.63KK2 pKa = 10.09IKK4 pKa = 10.66PITKK8 pKa = 9.17KK9 pKa = 10.51NGAIVYY15 pKa = 8.3RR16 pKa = 11.84ANVYY20 pKa = 10.65LGTDD24 pKa = 2.95QVTGKK29 pKa = 9.97KK30 pKa = 10.33AKK32 pKa = 9.51TSVTGRR38 pKa = 11.84TRR40 pKa = 11.84KK41 pKa = 8.76EE42 pKa = 3.84VKK44 pKa = 10.02QKK46 pKa = 10.96AKK48 pKa = 10.04HH49 pKa = 5.68AQDD52 pKa = 3.62EE53 pKa = 5.21FISNGYY59 pKa = 7.35TVTKK63 pKa = 9.88VVPIKK68 pKa = 10.67NYY70 pKa = 10.29QEE72 pKa = 4.68LAEE75 pKa = 4.74LWLEE79 pKa = 4.32SYY81 pKa = 10.85QLTVKK86 pKa = 9.85PQTFIATKK94 pKa = 10.24RR95 pKa = 11.84MLYY98 pKa = 10.01NHH100 pKa = 7.89LIPVFGALKK109 pKa = 10.32LNKK112 pKa = 10.01LSVSYY117 pKa = 10.64IQGFINDD124 pKa = 4.13LSAEE128 pKa = 4.15LVHH131 pKa = 6.38YY132 pKa = 10.48SVVHH136 pKa = 5.68SINRR140 pKa = 11.84RR141 pKa = 11.84VLQYY145 pKa = 10.88GVSLQLLPFNPARR158 pKa = 11.84DD159 pKa = 3.54VMLPKK164 pKa = 10.25VPKK167 pKa = 10.2KK168 pKa = 9.64EE169 pKa = 3.73NKK171 pKa = 9.48AIKK174 pKa = 10.16FIAPEE179 pKa = 3.97DD180 pKa = 3.88LKK182 pKa = 11.69ALMAYY187 pKa = 8.24MEE189 pKa = 4.47KK190 pKa = 10.52LANKK194 pKa = 9.07KK195 pKa = 9.8FSYY198 pKa = 10.51FFDD201 pKa = 3.67YY202 pKa = 11.08VLYY205 pKa = 10.68SVLLATGCRR214 pKa = 11.84FGEE217 pKa = 4.58VVALEE222 pKa = 4.12WSDD225 pKa = 3.32IDD227 pKa = 5.09LEE229 pKa = 4.52NGTISITKK237 pKa = 9.93NYY239 pKa = 10.09SRR241 pKa = 11.84LLKK244 pKa = 10.89LIGTPKK250 pKa = 9.91SKK252 pKa = 10.87AGVRR256 pKa = 11.84VISIDD261 pKa = 3.23KK262 pKa = 9.23KK263 pKa = 9.18TINLLRR269 pKa = 11.84LYY271 pKa = 10.58KK272 pKa = 10.31NRR274 pKa = 11.84QRR276 pKa = 11.84QIFIEE281 pKa = 4.08TGARR285 pKa = 11.84VSAVVFATPTKK296 pKa = 9.74EE297 pKa = 4.11YY298 pKa = 10.79QNMATRR304 pKa = 11.84QEE306 pKa = 4.21SLDD309 pKa = 3.62RR310 pKa = 11.84RR311 pKa = 11.84ITEE314 pKa = 3.41IGIPRR319 pKa = 11.84FTFHH323 pKa = 7.74AFRR326 pKa = 11.84HH327 pKa = 4.69THH329 pKa = 6.92ASLLLNAGISYY340 pKa = 10.68KK341 pKa = 10.23EE342 pKa = 3.61LQYY345 pKa = 11.49RR346 pKa = 11.84LGHH349 pKa = 6.19ATLAMTMDD357 pKa = 4.28IYY359 pKa = 11.69SHH361 pKa = 7.7LSMDD365 pKa = 3.69KK366 pKa = 10.11EE367 pKa = 4.32KK368 pKa = 10.95EE369 pKa = 3.93AVSYY373 pKa = 9.5YY374 pKa = 10.62EE375 pKa = 4.12KK376 pKa = 10.7AINSLL381 pKa = 3.57
Molecular weight: 43.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3076 |
37 |
507 |
146.5 |
16.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.722 ± 0.649 | 0.65 ± 0.202 |
6.404 ± 0.547 | 7.38 ± 0.497 |
3.771 ± 0.289 | 4.714 ± 0.376 |
1.658 ± 0.231 | 7.412 ± 0.373 |
8.713 ± 0.568 | 10.468 ± 0.49 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.568 ± 0.286 | 5.592 ± 0.346 |
3.771 ± 0.475 | 4.551 ± 0.398 |
4.876 ± 0.394 | 5.234 ± 0.372 |
6.014 ± 0.304 | 5.169 ± 0.481 |
0.943 ± 0.15 | 4.389 ± 0.36 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
