
Paraglaciecola arctica BSs20135
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Paraglaciecola; Paraglaciecola arctica
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
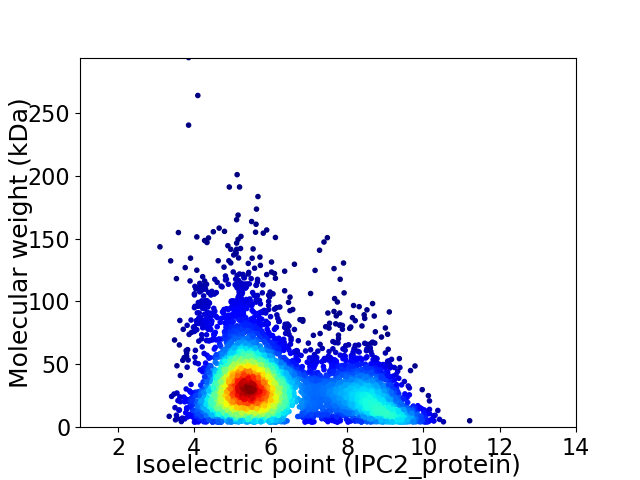
Virtual 2D-PAGE plot for 5330 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
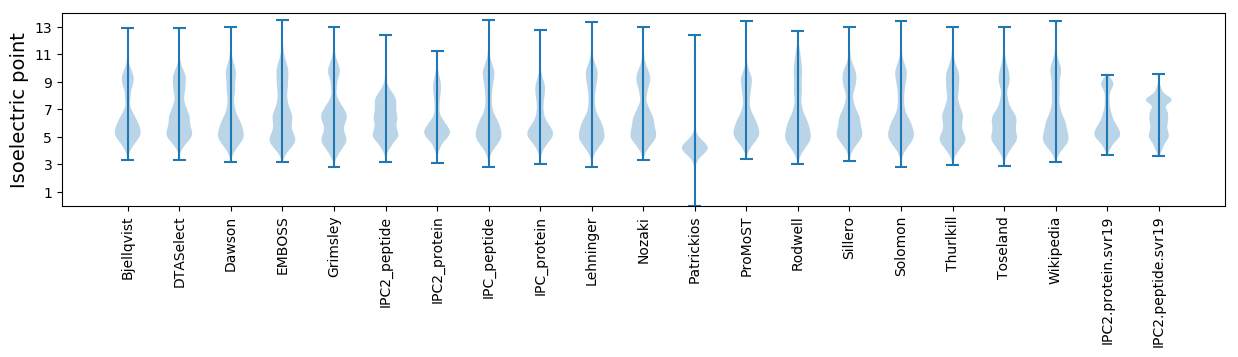
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K6ZE90|K6ZE90_9ALTE Uroporphyrinogen-III synthase OS=Paraglaciecola arctica BSs20135 OX=493475 GN=hemD PE=3 SV=1
MM1 pKa = 7.27LTLFKK6 pKa = 10.72KK7 pKa = 10.52SALTLSLITTLSILGGCGSDD27 pKa = 4.34SNDD30 pKa = 3.18APEE33 pKa = 4.29LSGNVLVTVAEE44 pKa = 4.22NTTEE48 pKa = 4.0VSQYY52 pKa = 10.02TATDD56 pKa = 3.61DD57 pKa = 4.04GTEE60 pKa = 3.96LTYY63 pKa = 10.8SITGSDD69 pKa = 3.65LQLFAIDD76 pKa = 3.38NTGTVEE82 pKa = 5.49FINAPDD88 pKa = 4.1FDD90 pKa = 4.82SGDD93 pKa = 3.42TGPYY97 pKa = 9.79NFTVTVTDD105 pKa = 4.16AEE107 pKa = 4.49GLTDD111 pKa = 4.95SIDD114 pKa = 3.4VTVNVGDD121 pKa = 4.13VLDD124 pKa = 4.39TPSLALVQTVAADD137 pKa = 3.7YY138 pKa = 10.99SSSQVAYY145 pKa = 10.75LNYY148 pKa = 10.45ASQTVEE154 pKa = 3.82DD155 pKa = 4.1GFYY158 pKa = 11.12AKK160 pKa = 9.04TQSDD164 pKa = 4.57YY165 pKa = 11.35SISIYY170 pKa = 10.56QSDD173 pKa = 4.55VYY175 pKa = 11.03HH176 pKa = 6.91IGRR179 pKa = 11.84YY180 pKa = 9.27FIDD183 pKa = 4.41TISKK187 pKa = 10.64YY188 pKa = 9.49NTDD191 pKa = 3.86DD192 pKa = 3.53ASTEE196 pKa = 3.37IWSYY200 pKa = 9.72STQDD204 pKa = 4.25DD205 pKa = 4.16LDD207 pKa = 3.79STTRR211 pKa = 11.84NPYY214 pKa = 9.23TLVSLSAEE222 pKa = 3.75KK223 pKa = 10.4AYY225 pKa = 10.78LLRR228 pKa = 11.84YY229 pKa = 9.88GSDD232 pKa = 3.66KK233 pKa = 10.62IWIVNPQATDD243 pKa = 3.73FEE245 pKa = 4.83SFKK248 pKa = 10.85IGEE251 pKa = 4.79LDD253 pKa = 3.52LASYY257 pKa = 11.04VPDD260 pKa = 3.76NNTSGTPRR268 pKa = 11.84PSAASINDD276 pKa = 3.1GKK278 pKa = 11.04LFVVMQRR285 pKa = 11.84SDD287 pKa = 3.44DD288 pKa = 4.15SYY290 pKa = 11.54GSTNSTYY297 pKa = 10.96LAVFDD302 pKa = 4.19TTTDD306 pKa = 3.63SEE308 pKa = 4.65IEE310 pKa = 4.27TNADD314 pKa = 3.07ATDD317 pKa = 3.71SVMGIPLLGLNPLEE331 pKa = 4.37HH332 pKa = 6.71SVIAFEE338 pKa = 4.52DD339 pKa = 3.29KK340 pKa = 11.17VYY342 pKa = 8.98ITSRR346 pKa = 11.84NSYY349 pKa = 8.27TAASVDD355 pKa = 3.26QSMIEE360 pKa = 3.9AVNTNDD366 pKa = 3.38YY367 pKa = 11.46SLTTVLSAADD377 pKa = 3.25ITDD380 pKa = 3.28NSTKK384 pKa = 10.38FIQSAAIVSAEE395 pKa = 3.68KK396 pKa = 10.79GYY398 pKa = 10.82FFANEE403 pKa = 4.14TFFTPYY409 pKa = 10.15HH410 pKa = 6.38EE411 pKa = 4.97EE412 pKa = 3.71STLYY416 pKa = 10.34QFNPTTGEE424 pKa = 3.73ISNEE428 pKa = 3.9TVLDD432 pKa = 3.47IDD434 pKa = 4.31SNSISFIDD442 pKa = 3.27IDD444 pKa = 3.58AAGFLWVSVKK454 pKa = 10.38EE455 pKa = 4.11DD456 pKa = 3.36AAPGVEE462 pKa = 5.2IIDD465 pKa = 3.97TKK467 pKa = 11.01TNTAVVTRR475 pKa = 11.84LEE477 pKa = 4.31TQLNPSVIRR486 pKa = 11.84FIEE489 pKa = 4.17EE490 pKa = 3.52
MM1 pKa = 7.27LTLFKK6 pKa = 10.72KK7 pKa = 10.52SALTLSLITTLSILGGCGSDD27 pKa = 4.34SNDD30 pKa = 3.18APEE33 pKa = 4.29LSGNVLVTVAEE44 pKa = 4.22NTTEE48 pKa = 4.0VSQYY52 pKa = 10.02TATDD56 pKa = 3.61DD57 pKa = 4.04GTEE60 pKa = 3.96LTYY63 pKa = 10.8SITGSDD69 pKa = 3.65LQLFAIDD76 pKa = 3.38NTGTVEE82 pKa = 5.49FINAPDD88 pKa = 4.1FDD90 pKa = 4.82SGDD93 pKa = 3.42TGPYY97 pKa = 9.79NFTVTVTDD105 pKa = 4.16AEE107 pKa = 4.49GLTDD111 pKa = 4.95SIDD114 pKa = 3.4VTVNVGDD121 pKa = 4.13VLDD124 pKa = 4.39TPSLALVQTVAADD137 pKa = 3.7YY138 pKa = 10.99SSSQVAYY145 pKa = 10.75LNYY148 pKa = 10.45ASQTVEE154 pKa = 3.82DD155 pKa = 4.1GFYY158 pKa = 11.12AKK160 pKa = 9.04TQSDD164 pKa = 4.57YY165 pKa = 11.35SISIYY170 pKa = 10.56QSDD173 pKa = 4.55VYY175 pKa = 11.03HH176 pKa = 6.91IGRR179 pKa = 11.84YY180 pKa = 9.27FIDD183 pKa = 4.41TISKK187 pKa = 10.64YY188 pKa = 9.49NTDD191 pKa = 3.86DD192 pKa = 3.53ASTEE196 pKa = 3.37IWSYY200 pKa = 9.72STQDD204 pKa = 4.25DD205 pKa = 4.16LDD207 pKa = 3.79STTRR211 pKa = 11.84NPYY214 pKa = 9.23TLVSLSAEE222 pKa = 3.75KK223 pKa = 10.4AYY225 pKa = 10.78LLRR228 pKa = 11.84YY229 pKa = 9.88GSDD232 pKa = 3.66KK233 pKa = 10.62IWIVNPQATDD243 pKa = 3.73FEE245 pKa = 4.83SFKK248 pKa = 10.85IGEE251 pKa = 4.79LDD253 pKa = 3.52LASYY257 pKa = 11.04VPDD260 pKa = 3.76NNTSGTPRR268 pKa = 11.84PSAASINDD276 pKa = 3.1GKK278 pKa = 11.04LFVVMQRR285 pKa = 11.84SDD287 pKa = 3.44DD288 pKa = 4.15SYY290 pKa = 11.54GSTNSTYY297 pKa = 10.96LAVFDD302 pKa = 4.19TTTDD306 pKa = 3.63SEE308 pKa = 4.65IEE310 pKa = 4.27TNADD314 pKa = 3.07ATDD317 pKa = 3.71SVMGIPLLGLNPLEE331 pKa = 4.37HH332 pKa = 6.71SVIAFEE338 pKa = 4.52DD339 pKa = 3.29KK340 pKa = 11.17VYY342 pKa = 8.98ITSRR346 pKa = 11.84NSYY349 pKa = 8.27TAASVDD355 pKa = 3.26QSMIEE360 pKa = 3.9AVNTNDD366 pKa = 3.38YY367 pKa = 11.46SLTTVLSAADD377 pKa = 3.25ITDD380 pKa = 3.28NSTKK384 pKa = 10.38FIQSAAIVSAEE395 pKa = 3.68KK396 pKa = 10.79GYY398 pKa = 10.82FFANEE403 pKa = 4.14TFFTPYY409 pKa = 10.15HH410 pKa = 6.38EE411 pKa = 4.97EE412 pKa = 3.71STLYY416 pKa = 10.34QFNPTTGEE424 pKa = 3.73ISNEE428 pKa = 3.9TVLDD432 pKa = 3.47IDD434 pKa = 4.31SNSISFIDD442 pKa = 3.27IDD444 pKa = 3.58AAGFLWVSVKK454 pKa = 10.38EE455 pKa = 4.11DD456 pKa = 3.36AAPGVEE462 pKa = 5.2IIDD465 pKa = 3.97TKK467 pKa = 11.01TNTAVVTRR475 pKa = 11.84LEE477 pKa = 4.31TQLNPSVIRR486 pKa = 11.84FIEE489 pKa = 4.17EE490 pKa = 3.52
Molecular weight: 53.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K6X8Y9|K6X8Y9_9ALTE Uncharacterized protein OS=Paraglaciecola arctica BSs20135 OX=493475 GN=GARC_0117 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 9.48VIANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 9.48VIANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1724099 |
37 |
2785 |
323.5 |
35.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.212 ± 0.037 | 0.977 ± 0.011 |
5.621 ± 0.037 | 5.837 ± 0.033 |
4.394 ± 0.023 | 6.619 ± 0.035 |
2.138 ± 0.018 | 6.821 ± 0.026 |
5.621 ± 0.03 | 10.404 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.46 ± 0.018 | 4.852 ± 0.029 |
3.746 ± 0.017 | 4.842 ± 0.035 |
3.965 ± 0.023 | 6.969 ± 0.031 |
5.444 ± 0.029 | 6.708 ± 0.028 |
1.245 ± 0.012 | 3.126 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
