
Klugiella xanthotipulae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Klugiella
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
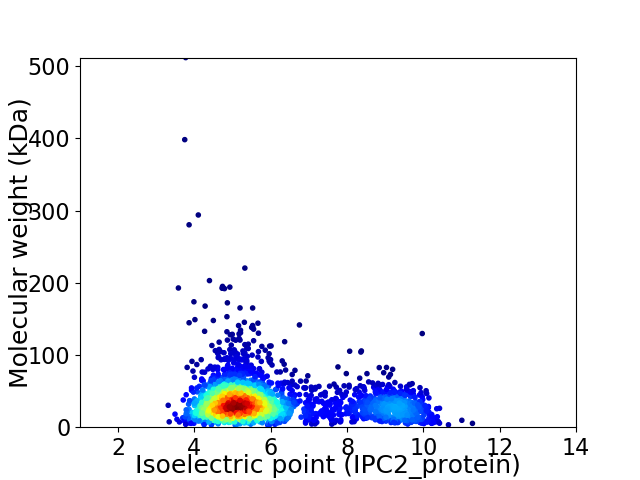
Virtual 2D-PAGE plot for 2573 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A543I682|A0A543I682_9MICO NadR type nicotinamide-nucleotide adenylyltransferase OS=Klugiella xanthotipulae OX=244735 GN=FB466_0909 PE=4 SV=1
MM1 pKa = 7.15LQVSGDD7 pKa = 3.49TSACRR12 pKa = 11.84PCNSGTDD19 pKa = 3.77LAQGGLTVHH28 pKa = 6.13NRR30 pKa = 11.84THH32 pKa = 7.34LAQHH36 pKa = 6.93HH37 pKa = 5.77GRR39 pKa = 11.84APATADD45 pKa = 3.27TPHH48 pKa = 7.43RR49 pKa = 11.84SRR51 pKa = 11.84SGSRR55 pKa = 11.84RR56 pKa = 11.84LTYY59 pKa = 9.55PALVAVLALGCGSLSLPAYY78 pKa = 8.72ATDD81 pKa = 3.83GDD83 pKa = 4.44VIDD86 pKa = 4.65GQSVAEE92 pKa = 4.38EE93 pKa = 4.22VQAAPPAADD102 pKa = 3.77TAPEE106 pKa = 4.09ALPVGEE112 pKa = 5.15AVPEE116 pKa = 4.37DD117 pKa = 3.84SAPAAVEE124 pKa = 4.25TPPVADD130 pKa = 3.84PVAVEE135 pKa = 4.81SPAEE139 pKa = 4.08APVAGPPVPVAAAAAAPSGITPFAGDD165 pKa = 4.08TILGSVSPATGDD177 pKa = 3.47EE178 pKa = 4.29AGGTTVILTGEE189 pKa = 4.45CFTDD193 pKa = 3.63AEE195 pKa = 4.25AVIFEE200 pKa = 4.53LTSAASFVVDD210 pKa = 4.18SDD212 pKa = 4.02TQITAVTPAGSGTVPVQVVGGPGCTFATLPNAFTYY247 pKa = 7.49EE248 pKa = 4.0APVPVVDD255 pKa = 4.4PPVVTSLTPDD265 pKa = 3.72RR266 pKa = 11.84GPVGGGSDD274 pKa = 3.49VTITGEE280 pKa = 3.9NLADD284 pKa = 3.66VTGVTVDD291 pKa = 4.72GIAATGLTGVTDD303 pKa = 3.87TSLTVTTPSHH313 pKa = 6.09AAGPVDD319 pKa = 3.77VVVTTTHH326 pKa = 5.48GTSNAGLFTYY336 pKa = 10.31HH337 pKa = 6.91PVASVTGVSPGSGSDD352 pKa = 3.24AGGTTVTITGTCFVGATGVTFGGTPAVSYY381 pKa = 11.08SVLTDD386 pKa = 3.27STLTAEE392 pKa = 4.68TPAGTGTVDD401 pKa = 3.32VSVATTGEE409 pKa = 4.37CGSGATLAGSFTYY422 pKa = 9.67TATVPAVPTITGISPTEE439 pKa = 4.26GPDD442 pKa = 3.2TGGTTVTVTGTNLDD456 pKa = 4.41GIQEE460 pKa = 4.2VTVDD464 pKa = 3.79GTAVAQFSLLTDD476 pKa = 3.79TSLTFVTEE484 pKa = 3.74AHH486 pKa = 6.51APGSVVVRR494 pKa = 11.84VRR496 pKa = 11.84DD497 pKa = 3.65ASDD500 pKa = 3.4EE501 pKa = 4.27SATSSFTFVAAPVVEE516 pKa = 4.83APAITSLSPTRR527 pKa = 11.84GTIDD531 pKa = 3.19GGTLVTITGEE541 pKa = 4.05RR542 pKa = 11.84LGAATAVAVGGTAVAAFTTVSDD564 pKa = 3.86TEE566 pKa = 4.27ITFEE570 pKa = 4.25TAAHH574 pKa = 6.35AAGTVDD580 pKa = 4.21VTVTDD585 pKa = 4.4ANGEE589 pKa = 4.19SAPAEE594 pKa = 4.04FTFFAPLAVTGILPATGSEE613 pKa = 4.14AGGEE617 pKa = 4.43TVTITGTCFTGAVDD631 pKa = 3.69VLFGSEE637 pKa = 4.0PATSFVVVTDD647 pKa = 3.57TSISAVVPAGSAGLVNVTVTGSVEE671 pKa = 4.12CGNSVLTGGYY681 pKa = 9.83EE682 pKa = 4.2YY683 pKa = 10.43VTVVQPVLGSITPDD697 pKa = 2.93SGPATGGTVVTATGSGFEE715 pKa = 4.29TAVSVTFDD723 pKa = 3.8GLPATNVTVVSDD735 pKa = 3.59TEE737 pKa = 4.21ITVVTPAHH745 pKa = 6.08GAGVVDD751 pKa = 3.87VVVNGPTDD759 pKa = 3.29SALQLSPFAVAFAGITPAASNAVSFTYY786 pKa = 10.33EE787 pKa = 3.67AVTTPGVGEE796 pKa = 4.19SGGAAGNVTGGAAGAPAPAPTAVGLAVTGLGGVTALLSAALLALVAGRR844 pKa = 11.84FLLVRR849 pKa = 11.84ARR851 pKa = 11.84RR852 pKa = 11.84GEE854 pKa = 3.91LSS856 pKa = 3.05
MM1 pKa = 7.15LQVSGDD7 pKa = 3.49TSACRR12 pKa = 11.84PCNSGTDD19 pKa = 3.77LAQGGLTVHH28 pKa = 6.13NRR30 pKa = 11.84THH32 pKa = 7.34LAQHH36 pKa = 6.93HH37 pKa = 5.77GRR39 pKa = 11.84APATADD45 pKa = 3.27TPHH48 pKa = 7.43RR49 pKa = 11.84SRR51 pKa = 11.84SGSRR55 pKa = 11.84RR56 pKa = 11.84LTYY59 pKa = 9.55PALVAVLALGCGSLSLPAYY78 pKa = 8.72ATDD81 pKa = 3.83GDD83 pKa = 4.44VIDD86 pKa = 4.65GQSVAEE92 pKa = 4.38EE93 pKa = 4.22VQAAPPAADD102 pKa = 3.77TAPEE106 pKa = 4.09ALPVGEE112 pKa = 5.15AVPEE116 pKa = 4.37DD117 pKa = 3.84SAPAAVEE124 pKa = 4.25TPPVADD130 pKa = 3.84PVAVEE135 pKa = 4.81SPAEE139 pKa = 4.08APVAGPPVPVAAAAAAPSGITPFAGDD165 pKa = 4.08TILGSVSPATGDD177 pKa = 3.47EE178 pKa = 4.29AGGTTVILTGEE189 pKa = 4.45CFTDD193 pKa = 3.63AEE195 pKa = 4.25AVIFEE200 pKa = 4.53LTSAASFVVDD210 pKa = 4.18SDD212 pKa = 4.02TQITAVTPAGSGTVPVQVVGGPGCTFATLPNAFTYY247 pKa = 7.49EE248 pKa = 4.0APVPVVDD255 pKa = 4.4PPVVTSLTPDD265 pKa = 3.72RR266 pKa = 11.84GPVGGGSDD274 pKa = 3.49VTITGEE280 pKa = 3.9NLADD284 pKa = 3.66VTGVTVDD291 pKa = 4.72GIAATGLTGVTDD303 pKa = 3.87TSLTVTTPSHH313 pKa = 6.09AAGPVDD319 pKa = 3.77VVVTTTHH326 pKa = 5.48GTSNAGLFTYY336 pKa = 10.31HH337 pKa = 6.91PVASVTGVSPGSGSDD352 pKa = 3.24AGGTTVTITGTCFVGATGVTFGGTPAVSYY381 pKa = 11.08SVLTDD386 pKa = 3.27STLTAEE392 pKa = 4.68TPAGTGTVDD401 pKa = 3.32VSVATTGEE409 pKa = 4.37CGSGATLAGSFTYY422 pKa = 9.67TATVPAVPTITGISPTEE439 pKa = 4.26GPDD442 pKa = 3.2TGGTTVTVTGTNLDD456 pKa = 4.41GIQEE460 pKa = 4.2VTVDD464 pKa = 3.79GTAVAQFSLLTDD476 pKa = 3.79TSLTFVTEE484 pKa = 3.74AHH486 pKa = 6.51APGSVVVRR494 pKa = 11.84VRR496 pKa = 11.84DD497 pKa = 3.65ASDD500 pKa = 3.4EE501 pKa = 4.27SATSSFTFVAAPVVEE516 pKa = 4.83APAITSLSPTRR527 pKa = 11.84GTIDD531 pKa = 3.19GGTLVTITGEE541 pKa = 4.05RR542 pKa = 11.84LGAATAVAVGGTAVAAFTTVSDD564 pKa = 3.86TEE566 pKa = 4.27ITFEE570 pKa = 4.25TAAHH574 pKa = 6.35AAGTVDD580 pKa = 4.21VTVTDD585 pKa = 4.4ANGEE589 pKa = 4.19SAPAEE594 pKa = 4.04FTFFAPLAVTGILPATGSEE613 pKa = 4.14AGGEE617 pKa = 4.43TVTITGTCFTGAVDD631 pKa = 3.69VLFGSEE637 pKa = 4.0PATSFVVVTDD647 pKa = 3.57TSISAVVPAGSAGLVNVTVTGSVEE671 pKa = 4.12CGNSVLTGGYY681 pKa = 9.83EE682 pKa = 4.2YY683 pKa = 10.43VTVVQPVLGSITPDD697 pKa = 2.93SGPATGGTVVTATGSGFEE715 pKa = 4.29TAVSVTFDD723 pKa = 3.8GLPATNVTVVSDD735 pKa = 3.59TEE737 pKa = 4.21ITVVTPAHH745 pKa = 6.08GAGVVDD751 pKa = 3.87VVVNGPTDD759 pKa = 3.29SALQLSPFAVAFAGITPAASNAVSFTYY786 pKa = 10.33EE787 pKa = 3.67AVTTPGVGEE796 pKa = 4.19SGGAAGNVTGGAAGAPAPAPTAVGLAVTGLGGVTALLSAALLALVAGRR844 pKa = 11.84FLLVRR849 pKa = 11.84ARR851 pKa = 11.84RR852 pKa = 11.84GEE854 pKa = 3.91LSS856 pKa = 3.05
Molecular weight: 82.81 kDa
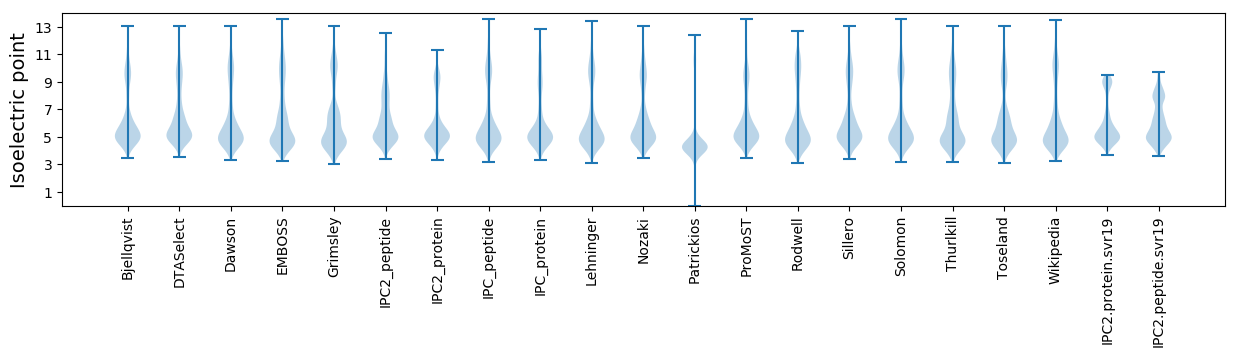
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A543I6V2|A0A543I6V2_9MICO DNA-binding response OmpR family regulator OS=Klugiella xanthotipulae OX=244735 GN=FB466_1118 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILSARR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.82GRR40 pKa = 11.84TKK42 pKa = 10.84LSAA45 pKa = 3.61
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILSARR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.82GRR40 pKa = 11.84TKK42 pKa = 10.84LSAA45 pKa = 3.61
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
880817 |
29 |
5270 |
342.3 |
36.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.104 ± 0.064 | 0.607 ± 0.014 |
5.61 ± 0.038 | 5.586 ± 0.052 |
3.104 ± 0.03 | 8.773 ± 0.068 |
2.0 ± 0.026 | 4.786 ± 0.038 |
2.098 ± 0.037 | 10.025 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.749 ± 0.023 | 2.587 ± 0.034 |
5.395 ± 0.04 | 2.9 ± 0.028 |
6.741 ± 0.07 | 6.435 ± 0.041 |
7.107 ± 0.091 | 8.82 ± 0.061 |
1.397 ± 0.024 | 2.175 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
