
Flavobacteriaceae bacterium Ap0902
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; unclassified Flavobacteriaceae
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
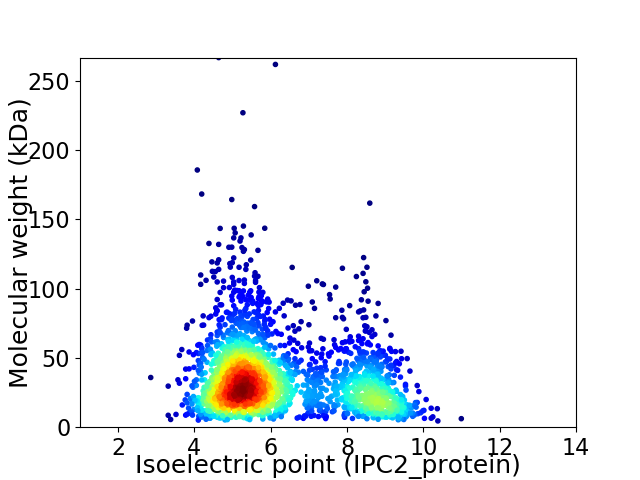
Virtual 2D-PAGE plot for 2350 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N8UHL2|A0A6N8UHL2_9FLAO 50S ribosomal protein L20 OS=Flavobacteriaceae bacterium Ap0902 OX=2684397 GN=rplT PE=3 SV=1
MM1 pKa = 8.09RR2 pKa = 11.84ITFTLLSAFCAILIQAQTVQDD23 pKa = 3.83AQKK26 pKa = 10.73KK27 pKa = 8.33IDD29 pKa = 3.68KK30 pKa = 10.88VYY32 pKa = 11.28DD33 pKa = 3.63NLNTSPYY40 pKa = 10.67LLQTEE45 pKa = 4.3QFKK48 pKa = 10.52IDD50 pKa = 4.13HH51 pKa = 6.25LHH53 pKa = 6.21SNPKK57 pKa = 9.75KK58 pKa = 10.63ALLNIQNPARR68 pKa = 11.84TEE70 pKa = 3.93QNIFTPLEE78 pKa = 4.34NKK80 pKa = 9.92PGDD83 pKa = 3.61YY84 pKa = 9.58WYY86 pKa = 11.43SSMDD90 pKa = 3.07WADD93 pKa = 3.2YY94 pKa = 11.08DD95 pKa = 4.15QDD97 pKa = 4.11GDD99 pKa = 4.21LDD101 pKa = 4.24LLISGAHH108 pKa = 5.65PRR110 pKa = 11.84EE111 pKa = 4.15GAPYY115 pKa = 10.06DD116 pKa = 3.72AGASTTHH123 pKa = 6.57VYY125 pKa = 11.26DD126 pKa = 3.54NDD128 pKa = 4.05GNGNLSLNTDD138 pKa = 3.94IILQGVHH145 pKa = 6.5ASDD148 pKa = 3.82VHH150 pKa = 6.59WLDD153 pKa = 3.42FDD155 pKa = 4.54YY156 pKa = 11.4DD157 pKa = 3.73GDD159 pKa = 4.38LDD161 pKa = 3.82IVMMGIDD168 pKa = 4.21YY169 pKa = 7.54DD170 pKa = 3.98TAFNMSALYY179 pKa = 10.21EE180 pKa = 4.31NNNGTFTAIEE190 pKa = 4.22VGTNGASWGGISIGDD205 pKa = 3.73YY206 pKa = 11.39DD207 pKa = 4.74NDD209 pKa = 3.84GDD211 pKa = 5.95LDD213 pKa = 3.75MLMTGWNVDD222 pKa = 4.39DD223 pKa = 4.95IDD225 pKa = 3.88NGYY228 pKa = 11.24ASTFLWEE235 pKa = 4.46NNDD238 pKa = 3.16GTFVRR243 pKa = 11.84NNISAQVPGVFNGNIAWVDD262 pKa = 3.0IDD264 pKa = 4.83NDD266 pKa = 3.42MDD268 pKa = 4.53LDD270 pKa = 3.96IVVSPAYY277 pKa = 10.17GVDD280 pKa = 3.49VQNLRR285 pKa = 11.84IYY287 pKa = 10.5KK288 pKa = 9.82RR289 pKa = 11.84EE290 pKa = 3.62NDD292 pKa = 3.8IYY294 pKa = 11.27TLTQKK299 pKa = 10.36VNNSSSYY306 pKa = 11.85AMFALGDD313 pKa = 3.78YY314 pKa = 10.94NNDD317 pKa = 3.03GFVDD321 pKa = 3.7IAVQTTDD328 pKa = 3.92DD329 pKa = 4.43DD330 pKa = 4.76YY331 pKa = 11.96NNFVDD336 pKa = 4.85ILKK339 pKa = 10.54NNQGDD344 pKa = 3.94FEE346 pKa = 4.63YY347 pKa = 11.21VEE349 pKa = 4.61SLPSVSSNGAVTPLAFGDD367 pKa = 3.85YY368 pKa = 11.02DD369 pKa = 4.16NDD371 pKa = 3.76GDD373 pKa = 5.38LDD375 pKa = 4.18LAVAGLDD382 pKa = 3.11SDD384 pKa = 4.73YY385 pKa = 11.3IGVSLLYY392 pKa = 10.54QNSDD396 pKa = 3.17NQFTIVNDD404 pKa = 3.53TGFDD408 pKa = 3.42KK409 pKa = 11.45VGGHH413 pKa = 5.43SLSWADD419 pKa = 3.7IDD421 pKa = 5.31GDD423 pKa = 4.2LDD425 pKa = 4.61LDD427 pKa = 3.64ILITGFIDD435 pKa = 4.94DD436 pKa = 3.97EE437 pKa = 5.43DD438 pKa = 3.8IGEE441 pKa = 4.37FYY443 pKa = 10.0GTSRR447 pKa = 11.84LYY449 pKa = 11.3KK450 pKa = 10.34NDD452 pKa = 3.11IGSANTAPNPPTTLSTQRR470 pKa = 11.84DD471 pKa = 3.61NGSVVFNWADD481 pKa = 3.16ASDD484 pKa = 3.92EE485 pKa = 4.04QTPTEE490 pKa = 4.06GLQYY494 pKa = 10.49QLQVGKK500 pKa = 10.62SPGGSDD506 pKa = 2.63IANYY510 pKa = 10.24SINNTSWVLNNLSPEE525 pKa = 4.1TTYY528 pKa = 10.67YY529 pKa = 9.94WCVKK533 pKa = 10.56SIDD536 pKa = 3.57TSFVKK541 pKa = 10.26SQCSEE546 pKa = 3.64EE547 pKa = 4.12QMISNLSTEE556 pKa = 4.42EE557 pKa = 3.91VNLDD561 pKa = 3.59KK562 pKa = 11.45NVVRR566 pKa = 11.84LVSNPVEE573 pKa = 3.85NGIIHH578 pKa = 7.24LVTSEE583 pKa = 3.74WGHH586 pKa = 7.42PEE588 pKa = 4.0MQVKK592 pKa = 10.18LYY594 pKa = 10.98DD595 pKa = 3.64MKK597 pKa = 11.31GGLILNKK604 pKa = 9.33TFNDD608 pKa = 2.76IRR610 pKa = 11.84NIIDD614 pKa = 3.47IPVHH618 pKa = 5.77HH619 pKa = 7.44LSTGKK624 pKa = 10.86YY625 pKa = 8.29ILQYY629 pKa = 10.83QNQTDD634 pKa = 3.78TGALPIIIKK643 pKa = 9.42QQ644 pKa = 3.3
MM1 pKa = 8.09RR2 pKa = 11.84ITFTLLSAFCAILIQAQTVQDD23 pKa = 3.83AQKK26 pKa = 10.73KK27 pKa = 8.33IDD29 pKa = 3.68KK30 pKa = 10.88VYY32 pKa = 11.28DD33 pKa = 3.63NLNTSPYY40 pKa = 10.67LLQTEE45 pKa = 4.3QFKK48 pKa = 10.52IDD50 pKa = 4.13HH51 pKa = 6.25LHH53 pKa = 6.21SNPKK57 pKa = 9.75KK58 pKa = 10.63ALLNIQNPARR68 pKa = 11.84TEE70 pKa = 3.93QNIFTPLEE78 pKa = 4.34NKK80 pKa = 9.92PGDD83 pKa = 3.61YY84 pKa = 9.58WYY86 pKa = 11.43SSMDD90 pKa = 3.07WADD93 pKa = 3.2YY94 pKa = 11.08DD95 pKa = 4.15QDD97 pKa = 4.11GDD99 pKa = 4.21LDD101 pKa = 4.24LLISGAHH108 pKa = 5.65PRR110 pKa = 11.84EE111 pKa = 4.15GAPYY115 pKa = 10.06DD116 pKa = 3.72AGASTTHH123 pKa = 6.57VYY125 pKa = 11.26DD126 pKa = 3.54NDD128 pKa = 4.05GNGNLSLNTDD138 pKa = 3.94IILQGVHH145 pKa = 6.5ASDD148 pKa = 3.82VHH150 pKa = 6.59WLDD153 pKa = 3.42FDD155 pKa = 4.54YY156 pKa = 11.4DD157 pKa = 3.73GDD159 pKa = 4.38LDD161 pKa = 3.82IVMMGIDD168 pKa = 4.21YY169 pKa = 7.54DD170 pKa = 3.98TAFNMSALYY179 pKa = 10.21EE180 pKa = 4.31NNNGTFTAIEE190 pKa = 4.22VGTNGASWGGISIGDD205 pKa = 3.73YY206 pKa = 11.39DD207 pKa = 4.74NDD209 pKa = 3.84GDD211 pKa = 5.95LDD213 pKa = 3.75MLMTGWNVDD222 pKa = 4.39DD223 pKa = 4.95IDD225 pKa = 3.88NGYY228 pKa = 11.24ASTFLWEE235 pKa = 4.46NNDD238 pKa = 3.16GTFVRR243 pKa = 11.84NNISAQVPGVFNGNIAWVDD262 pKa = 3.0IDD264 pKa = 4.83NDD266 pKa = 3.42MDD268 pKa = 4.53LDD270 pKa = 3.96IVVSPAYY277 pKa = 10.17GVDD280 pKa = 3.49VQNLRR285 pKa = 11.84IYY287 pKa = 10.5KK288 pKa = 9.82RR289 pKa = 11.84EE290 pKa = 3.62NDD292 pKa = 3.8IYY294 pKa = 11.27TLTQKK299 pKa = 10.36VNNSSSYY306 pKa = 11.85AMFALGDD313 pKa = 3.78YY314 pKa = 10.94NNDD317 pKa = 3.03GFVDD321 pKa = 3.7IAVQTTDD328 pKa = 3.92DD329 pKa = 4.43DD330 pKa = 4.76YY331 pKa = 11.96NNFVDD336 pKa = 4.85ILKK339 pKa = 10.54NNQGDD344 pKa = 3.94FEE346 pKa = 4.63YY347 pKa = 11.21VEE349 pKa = 4.61SLPSVSSNGAVTPLAFGDD367 pKa = 3.85YY368 pKa = 11.02DD369 pKa = 4.16NDD371 pKa = 3.76GDD373 pKa = 5.38LDD375 pKa = 4.18LAVAGLDD382 pKa = 3.11SDD384 pKa = 4.73YY385 pKa = 11.3IGVSLLYY392 pKa = 10.54QNSDD396 pKa = 3.17NQFTIVNDD404 pKa = 3.53TGFDD408 pKa = 3.42KK409 pKa = 11.45VGGHH413 pKa = 5.43SLSWADD419 pKa = 3.7IDD421 pKa = 5.31GDD423 pKa = 4.2LDD425 pKa = 4.61LDD427 pKa = 3.64ILITGFIDD435 pKa = 4.94DD436 pKa = 3.97EE437 pKa = 5.43DD438 pKa = 3.8IGEE441 pKa = 4.37FYY443 pKa = 10.0GTSRR447 pKa = 11.84LYY449 pKa = 11.3KK450 pKa = 10.34NDD452 pKa = 3.11IGSANTAPNPPTTLSTQRR470 pKa = 11.84DD471 pKa = 3.61NGSVVFNWADD481 pKa = 3.16ASDD484 pKa = 3.92EE485 pKa = 4.04QTPTEE490 pKa = 4.06GLQYY494 pKa = 10.49QLQVGKK500 pKa = 10.62SPGGSDD506 pKa = 2.63IANYY510 pKa = 10.24SINNTSWVLNNLSPEE525 pKa = 4.1TTYY528 pKa = 10.67YY529 pKa = 9.94WCVKK533 pKa = 10.56SIDD536 pKa = 3.57TSFVKK541 pKa = 10.26SQCSEE546 pKa = 3.64EE547 pKa = 4.12QMISNLSTEE556 pKa = 4.42EE557 pKa = 3.91VNLDD561 pKa = 3.59KK562 pKa = 11.45NVVRR566 pKa = 11.84LVSNPVEE573 pKa = 3.85NGIIHH578 pKa = 7.24LVTSEE583 pKa = 3.74WGHH586 pKa = 7.42PEE588 pKa = 4.0MQVKK592 pKa = 10.18LYY594 pKa = 10.98DD595 pKa = 3.64MKK597 pKa = 11.31GGLILNKK604 pKa = 9.33TFNDD608 pKa = 2.76IRR610 pKa = 11.84NIIDD614 pKa = 3.47IPVHH618 pKa = 5.77HH619 pKa = 7.44LSTGKK624 pKa = 10.86YY625 pKa = 8.29ILQYY629 pKa = 10.83QNQTDD634 pKa = 3.78TGALPIIIKK643 pKa = 9.42QQ644 pKa = 3.3
Molecular weight: 71.4 kDa
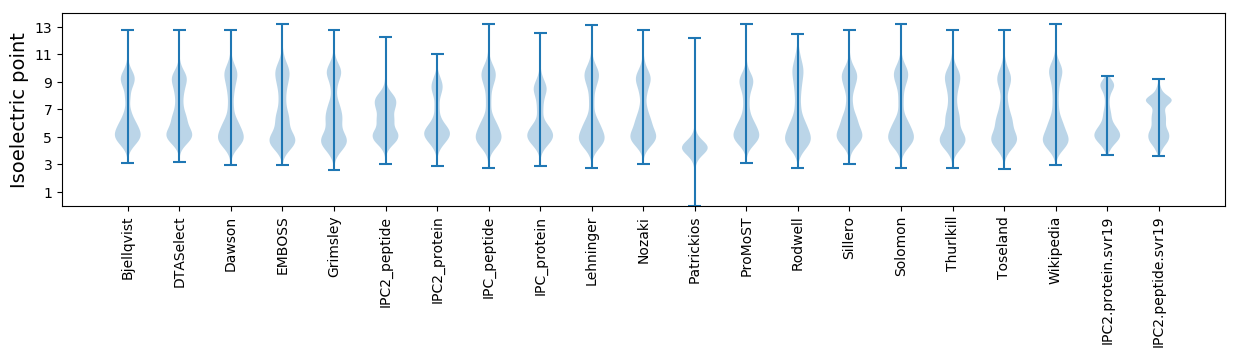
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N8UGE4|A0A6N8UGE4_9FLAO 50S ribosomal protein L4 OS=Flavobacteriaceae bacterium Ap0902 OX=2684397 GN=rplD PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.86KK12 pKa = 10.03RR13 pKa = 11.84NKK15 pKa = 9.59HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.1RR21 pKa = 11.84MSTKK25 pKa = 9.7NGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.65GRR39 pKa = 11.84KK40 pKa = 8.69NISVSTVRR48 pKa = 11.84AKK50 pKa = 10.57RR51 pKa = 3.36
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.86KK12 pKa = 10.03RR13 pKa = 11.84NKK15 pKa = 9.59HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.1RR21 pKa = 11.84MSTKK25 pKa = 9.7NGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.65GRR39 pKa = 11.84KK40 pKa = 8.69NISVSTVRR48 pKa = 11.84AKK50 pKa = 10.57RR51 pKa = 3.36
Molecular weight: 6.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
768563 |
38 |
2351 |
327.0 |
37.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.326 ± 0.052 | 0.617 ± 0.015 |
5.85 ± 0.037 | 6.979 ± 0.055 |
5.028 ± 0.041 | 6.373 ± 0.056 |
1.895 ± 0.023 | 8.125 ± 0.051 |
7.521 ± 0.059 | 9.276 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.433 ± 0.025 | 6.086 ± 0.048 |
3.462 ± 0.028 | 3.702 ± 0.028 |
3.623 ± 0.03 | 6.002 ± 0.035 |
5.321 ± 0.031 | 5.996 ± 0.036 |
1.083 ± 0.02 | 4.301 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
