
Wenzhou yanvirus-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.35
Get precalculated fractions of proteins
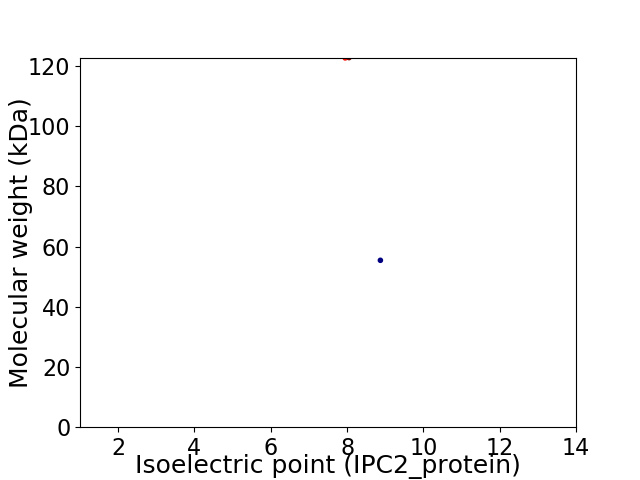
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
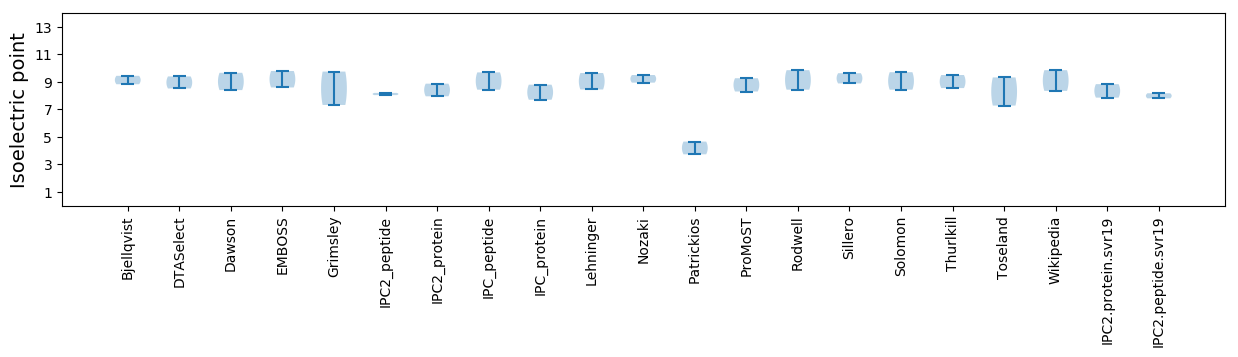
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL64|A0A1L3KL64_9VIRU RNA-dependent RNA polymerase OS=Wenzhou yanvirus-like virus 1 OX=1923683 PE=4 SV=1
MM1 pKa = 7.71VDD3 pKa = 2.92QCRR6 pKa = 11.84FLGLSDD12 pKa = 3.53FPIRR16 pKa = 11.84VMGPVGTVSHH26 pKa = 5.74YY27 pKa = 10.8VCGKK31 pKa = 8.86PRR33 pKa = 11.84IDD35 pKa = 3.98GACLLYY41 pKa = 10.44IPARR45 pKa = 11.84PPLVPLAHH53 pKa = 6.0WTIAVGGQSYY63 pKa = 10.87DD64 pKa = 3.69YY65 pKa = 10.93ADD67 pKa = 3.97PAALGGVVLYY77 pKa = 11.07SDD79 pKa = 4.98IPPISYY85 pKa = 9.64NPHH88 pKa = 5.18VFWHH92 pKa = 6.51AGSCKK97 pKa = 10.23SATLQFWMRR106 pKa = 11.84AGNGCRR112 pKa = 11.84CKK114 pKa = 10.4RR115 pKa = 11.84FCRR118 pKa = 11.84HH119 pKa = 4.35QFMWEE124 pKa = 4.12YY125 pKa = 10.84NHH127 pKa = 6.54EE128 pKa = 4.1LANRR132 pKa = 11.84GLPAGTGPVAFMLSCRR148 pKa = 11.84IHH150 pKa = 6.19PRR152 pKa = 11.84KK153 pKa = 9.93LYY155 pKa = 10.3NPRR158 pKa = 11.84TVRR161 pKa = 11.84FGSGRR166 pKa = 11.84PGQYY170 pKa = 8.51VAHH173 pKa = 7.75ILRR176 pKa = 11.84CPYY179 pKa = 10.05SDD181 pKa = 3.55YY182 pKa = 11.36DD183 pKa = 3.82VRR185 pKa = 11.84ISNCTTVQIVTSRR198 pKa = 11.84GIVYY202 pKa = 9.5SQNKK206 pKa = 6.48ATSMEE211 pKa = 4.46FLNKK215 pKa = 8.6THH217 pKa = 6.85LCVPTGEE224 pKa = 3.95FMQWGGVALEE234 pKa = 4.41LLRR237 pKa = 11.84IEE239 pKa = 4.29PQPVISFTPSPRR251 pKa = 11.84TRR253 pKa = 11.84GRR255 pKa = 11.84FYY257 pKa = 11.21LVVGVAKK264 pKa = 10.21AVSSMFSRR272 pKa = 11.84NPLVRR277 pKa = 11.84YY278 pKa = 8.61HH279 pKa = 3.92VTYY282 pKa = 10.69RR283 pKa = 11.84RR284 pKa = 11.84FASASTFCAIGVAGALMFEE303 pKa = 4.64LRR305 pKa = 11.84SIARR309 pKa = 11.84SLFNMRR315 pKa = 11.84SQQRR319 pKa = 11.84FFPMQVEE326 pKa = 4.15YY327 pKa = 10.97DD328 pKa = 3.72VATFAPYY335 pKa = 10.38NIQLPFEE342 pKa = 4.28QEE344 pKa = 3.47LFMRR348 pKa = 11.84MAVKK352 pKa = 10.31RR353 pKa = 11.84DD354 pKa = 3.15ITDD357 pKa = 2.4AWIRR361 pKa = 11.84DD362 pKa = 3.88VITRR366 pKa = 11.84LAHH369 pKa = 5.15QNDD372 pKa = 3.11WRR374 pKa = 11.84GVLNRR379 pKa = 11.84HH380 pKa = 6.27EE381 pKa = 4.36LNQWISRR388 pKa = 11.84VVTEE392 pKa = 5.21PGLVWEE398 pKa = 4.41VQKK401 pKa = 11.39DD402 pKa = 3.78GLRR405 pKa = 11.84NNACYY410 pKa = 9.45TCLSIGRR417 pKa = 11.84KK418 pKa = 6.85MRR420 pKa = 11.84RR421 pKa = 11.84NEE423 pKa = 3.82CAQCRR428 pKa = 11.84TQRR431 pKa = 11.84NLSPLYY437 pKa = 7.53FTPIPYY443 pKa = 9.27TPFAHH448 pKa = 6.11VGLVGIYY455 pKa = 9.84SRR457 pKa = 11.84PPLLPPATYY466 pKa = 10.23YY467 pKa = 10.53HH468 pKa = 6.24SCEE471 pKa = 4.06EE472 pKa = 4.08RR473 pKa = 11.84YY474 pKa = 10.11LPYY477 pKa = 10.44KK478 pKa = 10.22SGGWYY483 pKa = 9.29PGRR486 pKa = 11.84PFEE489 pKa = 5.16FIYY492 pKa = 10.48HH493 pKa = 6.59CSYY496 pKa = 10.15KK497 pKa = 10.82GKK499 pKa = 10.11IVNDD503 pKa = 3.87PEE505 pKa = 3.97TAYY508 pKa = 10.45AIYY511 pKa = 10.69NSFGPQPRR519 pKa = 11.84CRR521 pKa = 11.84GKK523 pKa = 10.81LCGPMLAGAHH533 pKa = 5.17MQCFTSGTEE542 pKa = 3.83TALLAFVGRR551 pKa = 11.84MGAVSPEE558 pKa = 3.92YY559 pKa = 10.34QHH561 pKa = 6.42YY562 pKa = 10.78LSLGVNHH569 pKa = 6.35SVYY572 pKa = 11.23NDD574 pKa = 3.16IAKK577 pKa = 10.47VNARR581 pKa = 11.84HH582 pKa = 6.02KK583 pKa = 10.55FDD585 pKa = 3.73YY586 pKa = 10.74LLEE589 pKa = 4.34IFKK592 pKa = 10.86HH593 pKa = 5.27VGAAAYY599 pKa = 10.72YY600 pKa = 10.82DD601 pKa = 3.87GDD603 pKa = 3.77NDD605 pKa = 4.71AGGLRR610 pKa = 11.84SKK612 pKa = 10.1YY613 pKa = 8.72IPSRR617 pKa = 11.84EE618 pKa = 4.07VVPWTPQQVIDD629 pKa = 4.54HH630 pKa = 6.31QPDD633 pKa = 3.33SAKK636 pKa = 10.17RR637 pKa = 11.84RR638 pKa = 11.84IMIEE642 pKa = 3.83AYY644 pKa = 10.21RR645 pKa = 11.84DD646 pKa = 3.15IDD648 pKa = 3.92EE649 pKa = 5.48GFLTPTPKK657 pKa = 10.4LYY659 pKa = 10.06KK660 pKa = 10.34AKK662 pKa = 10.57AIAKK666 pKa = 9.26GEE668 pKa = 3.82KK669 pKa = 10.09SMAWDD674 pKa = 3.5HH675 pKa = 7.15DD676 pKa = 3.58ISEE679 pKa = 4.27TLIYY683 pKa = 10.17KK684 pKa = 10.02SKK686 pKa = 10.2QVPRR690 pKa = 11.84FISACSPHH698 pKa = 6.34TNALIAPYY706 pKa = 9.57IAAAEE711 pKa = 4.12EE712 pKa = 4.37FVKK715 pKa = 10.22EE716 pKa = 4.36FCSSSASVAYY726 pKa = 10.09ASGMNPEE733 pKa = 4.91QLNEE737 pKa = 3.96WLNTYY742 pKa = 10.55AVDD745 pKa = 3.74RR746 pKa = 11.84LIIEE750 pKa = 4.61GDD752 pKa = 3.12ISSADD757 pKa = 3.39SAIGVEE763 pKa = 3.79AHH765 pKa = 6.31YY766 pKa = 11.04LFFYY770 pKa = 10.6FLTWLFPEE778 pKa = 4.95LPEE781 pKa = 4.97SDD783 pKa = 3.37VWHH786 pKa = 6.63VIQALSYY793 pKa = 10.4IYY795 pKa = 10.66VDD797 pKa = 3.27RR798 pKa = 11.84DD799 pKa = 3.03GSKK802 pKa = 10.49FVAGPVNVSGSRR814 pKa = 11.84YY815 pKa = 6.98TSCIVTIWVMVANVTSSAFSCLSDD839 pKa = 3.83YY840 pKa = 9.25RR841 pKa = 11.84TLSQADD847 pKa = 4.29RR848 pKa = 11.84IGMVKK853 pKa = 10.5GLLQSGNAATITCSDD868 pKa = 4.69DD869 pKa = 3.0IYY871 pKa = 10.68TAVNTSFSEE880 pKa = 3.75WHH882 pKa = 5.7FMRR885 pKa = 11.84NISPYY890 pKa = 9.89LRR892 pKa = 11.84AYY894 pKa = 9.37TYY896 pKa = 9.92EE897 pKa = 4.42CSVDD901 pKa = 3.93SAWLRR906 pKa = 11.84RR907 pKa = 11.84FCDD910 pKa = 3.28TMRR913 pKa = 11.84TEE915 pKa = 4.37FFLDD919 pKa = 3.26VRR921 pKa = 11.84PNKK924 pKa = 9.89CRR926 pKa = 11.84LFPAAEE932 pKa = 3.64WRR934 pKa = 11.84LATFLAMRR942 pKa = 11.84PVWSGSRR949 pKa = 11.84YY950 pKa = 7.5EE951 pKa = 4.34WGPEE955 pKa = 2.86ICRR958 pKa = 11.84RR959 pKa = 11.84MRR961 pKa = 11.84SAYY964 pKa = 8.71WMFDD968 pKa = 3.15KK969 pKa = 10.48EE970 pKa = 4.7HH971 pKa = 7.14PPMAWGRR978 pKa = 11.84GISAALLVAGRR989 pKa = 11.84HH990 pKa = 4.53VPVLRR995 pKa = 11.84DD996 pKa = 2.77ICTWHH1001 pKa = 6.98LSVTHH1006 pKa = 6.07GPIMKK1011 pKa = 10.36LPFTNIWSTFYY1022 pKa = 10.32RR1023 pKa = 11.84YY1024 pKa = 9.01EE1025 pKa = 3.91VSGEE1029 pKa = 3.89FTDD1032 pKa = 4.31RR1033 pKa = 11.84GLNEE1037 pKa = 3.96FLADD1041 pKa = 3.8YY1042 pKa = 10.36DD1043 pKa = 3.82ISTSEE1048 pKa = 4.0YY1049 pKa = 10.66LDD1051 pKa = 4.09FVDD1054 pKa = 5.38KK1055 pKa = 11.3LKK1057 pKa = 11.38GCMDD1061 pKa = 4.27PFINIQHH1068 pKa = 5.86SVIQKK1073 pKa = 10.15LLMRR1077 pKa = 11.84EE1078 pKa = 4.05
MM1 pKa = 7.71VDD3 pKa = 2.92QCRR6 pKa = 11.84FLGLSDD12 pKa = 3.53FPIRR16 pKa = 11.84VMGPVGTVSHH26 pKa = 5.74YY27 pKa = 10.8VCGKK31 pKa = 8.86PRR33 pKa = 11.84IDD35 pKa = 3.98GACLLYY41 pKa = 10.44IPARR45 pKa = 11.84PPLVPLAHH53 pKa = 6.0WTIAVGGQSYY63 pKa = 10.87DD64 pKa = 3.69YY65 pKa = 10.93ADD67 pKa = 3.97PAALGGVVLYY77 pKa = 11.07SDD79 pKa = 4.98IPPISYY85 pKa = 9.64NPHH88 pKa = 5.18VFWHH92 pKa = 6.51AGSCKK97 pKa = 10.23SATLQFWMRR106 pKa = 11.84AGNGCRR112 pKa = 11.84CKK114 pKa = 10.4RR115 pKa = 11.84FCRR118 pKa = 11.84HH119 pKa = 4.35QFMWEE124 pKa = 4.12YY125 pKa = 10.84NHH127 pKa = 6.54EE128 pKa = 4.1LANRR132 pKa = 11.84GLPAGTGPVAFMLSCRR148 pKa = 11.84IHH150 pKa = 6.19PRR152 pKa = 11.84KK153 pKa = 9.93LYY155 pKa = 10.3NPRR158 pKa = 11.84TVRR161 pKa = 11.84FGSGRR166 pKa = 11.84PGQYY170 pKa = 8.51VAHH173 pKa = 7.75ILRR176 pKa = 11.84CPYY179 pKa = 10.05SDD181 pKa = 3.55YY182 pKa = 11.36DD183 pKa = 3.82VRR185 pKa = 11.84ISNCTTVQIVTSRR198 pKa = 11.84GIVYY202 pKa = 9.5SQNKK206 pKa = 6.48ATSMEE211 pKa = 4.46FLNKK215 pKa = 8.6THH217 pKa = 6.85LCVPTGEE224 pKa = 3.95FMQWGGVALEE234 pKa = 4.41LLRR237 pKa = 11.84IEE239 pKa = 4.29PQPVISFTPSPRR251 pKa = 11.84TRR253 pKa = 11.84GRR255 pKa = 11.84FYY257 pKa = 11.21LVVGVAKK264 pKa = 10.21AVSSMFSRR272 pKa = 11.84NPLVRR277 pKa = 11.84YY278 pKa = 8.61HH279 pKa = 3.92VTYY282 pKa = 10.69RR283 pKa = 11.84RR284 pKa = 11.84FASASTFCAIGVAGALMFEE303 pKa = 4.64LRR305 pKa = 11.84SIARR309 pKa = 11.84SLFNMRR315 pKa = 11.84SQQRR319 pKa = 11.84FFPMQVEE326 pKa = 4.15YY327 pKa = 10.97DD328 pKa = 3.72VATFAPYY335 pKa = 10.38NIQLPFEE342 pKa = 4.28QEE344 pKa = 3.47LFMRR348 pKa = 11.84MAVKK352 pKa = 10.31RR353 pKa = 11.84DD354 pKa = 3.15ITDD357 pKa = 2.4AWIRR361 pKa = 11.84DD362 pKa = 3.88VITRR366 pKa = 11.84LAHH369 pKa = 5.15QNDD372 pKa = 3.11WRR374 pKa = 11.84GVLNRR379 pKa = 11.84HH380 pKa = 6.27EE381 pKa = 4.36LNQWISRR388 pKa = 11.84VVTEE392 pKa = 5.21PGLVWEE398 pKa = 4.41VQKK401 pKa = 11.39DD402 pKa = 3.78GLRR405 pKa = 11.84NNACYY410 pKa = 9.45TCLSIGRR417 pKa = 11.84KK418 pKa = 6.85MRR420 pKa = 11.84RR421 pKa = 11.84NEE423 pKa = 3.82CAQCRR428 pKa = 11.84TQRR431 pKa = 11.84NLSPLYY437 pKa = 7.53FTPIPYY443 pKa = 9.27TPFAHH448 pKa = 6.11VGLVGIYY455 pKa = 9.84SRR457 pKa = 11.84PPLLPPATYY466 pKa = 10.23YY467 pKa = 10.53HH468 pKa = 6.24SCEE471 pKa = 4.06EE472 pKa = 4.08RR473 pKa = 11.84YY474 pKa = 10.11LPYY477 pKa = 10.44KK478 pKa = 10.22SGGWYY483 pKa = 9.29PGRR486 pKa = 11.84PFEE489 pKa = 5.16FIYY492 pKa = 10.48HH493 pKa = 6.59CSYY496 pKa = 10.15KK497 pKa = 10.82GKK499 pKa = 10.11IVNDD503 pKa = 3.87PEE505 pKa = 3.97TAYY508 pKa = 10.45AIYY511 pKa = 10.69NSFGPQPRR519 pKa = 11.84CRR521 pKa = 11.84GKK523 pKa = 10.81LCGPMLAGAHH533 pKa = 5.17MQCFTSGTEE542 pKa = 3.83TALLAFVGRR551 pKa = 11.84MGAVSPEE558 pKa = 3.92YY559 pKa = 10.34QHH561 pKa = 6.42YY562 pKa = 10.78LSLGVNHH569 pKa = 6.35SVYY572 pKa = 11.23NDD574 pKa = 3.16IAKK577 pKa = 10.47VNARR581 pKa = 11.84HH582 pKa = 6.02KK583 pKa = 10.55FDD585 pKa = 3.73YY586 pKa = 10.74LLEE589 pKa = 4.34IFKK592 pKa = 10.86HH593 pKa = 5.27VGAAAYY599 pKa = 10.72YY600 pKa = 10.82DD601 pKa = 3.87GDD603 pKa = 3.77NDD605 pKa = 4.71AGGLRR610 pKa = 11.84SKK612 pKa = 10.1YY613 pKa = 8.72IPSRR617 pKa = 11.84EE618 pKa = 4.07VVPWTPQQVIDD629 pKa = 4.54HH630 pKa = 6.31QPDD633 pKa = 3.33SAKK636 pKa = 10.17RR637 pKa = 11.84RR638 pKa = 11.84IMIEE642 pKa = 3.83AYY644 pKa = 10.21RR645 pKa = 11.84DD646 pKa = 3.15IDD648 pKa = 3.92EE649 pKa = 5.48GFLTPTPKK657 pKa = 10.4LYY659 pKa = 10.06KK660 pKa = 10.34AKK662 pKa = 10.57AIAKK666 pKa = 9.26GEE668 pKa = 3.82KK669 pKa = 10.09SMAWDD674 pKa = 3.5HH675 pKa = 7.15DD676 pKa = 3.58ISEE679 pKa = 4.27TLIYY683 pKa = 10.17KK684 pKa = 10.02SKK686 pKa = 10.2QVPRR690 pKa = 11.84FISACSPHH698 pKa = 6.34TNALIAPYY706 pKa = 9.57IAAAEE711 pKa = 4.12EE712 pKa = 4.37FVKK715 pKa = 10.22EE716 pKa = 4.36FCSSSASVAYY726 pKa = 10.09ASGMNPEE733 pKa = 4.91QLNEE737 pKa = 3.96WLNTYY742 pKa = 10.55AVDD745 pKa = 3.74RR746 pKa = 11.84LIIEE750 pKa = 4.61GDD752 pKa = 3.12ISSADD757 pKa = 3.39SAIGVEE763 pKa = 3.79AHH765 pKa = 6.31YY766 pKa = 11.04LFFYY770 pKa = 10.6FLTWLFPEE778 pKa = 4.95LPEE781 pKa = 4.97SDD783 pKa = 3.37VWHH786 pKa = 6.63VIQALSYY793 pKa = 10.4IYY795 pKa = 10.66VDD797 pKa = 3.27RR798 pKa = 11.84DD799 pKa = 3.03GSKK802 pKa = 10.49FVAGPVNVSGSRR814 pKa = 11.84YY815 pKa = 6.98TSCIVTIWVMVANVTSSAFSCLSDD839 pKa = 3.83YY840 pKa = 9.25RR841 pKa = 11.84TLSQADD847 pKa = 4.29RR848 pKa = 11.84IGMVKK853 pKa = 10.5GLLQSGNAATITCSDD868 pKa = 4.69DD869 pKa = 3.0IYY871 pKa = 10.68TAVNTSFSEE880 pKa = 3.75WHH882 pKa = 5.7FMRR885 pKa = 11.84NISPYY890 pKa = 9.89LRR892 pKa = 11.84AYY894 pKa = 9.37TYY896 pKa = 9.92EE897 pKa = 4.42CSVDD901 pKa = 3.93SAWLRR906 pKa = 11.84RR907 pKa = 11.84FCDD910 pKa = 3.28TMRR913 pKa = 11.84TEE915 pKa = 4.37FFLDD919 pKa = 3.26VRR921 pKa = 11.84PNKK924 pKa = 9.89CRR926 pKa = 11.84LFPAAEE932 pKa = 3.64WRR934 pKa = 11.84LATFLAMRR942 pKa = 11.84PVWSGSRR949 pKa = 11.84YY950 pKa = 7.5EE951 pKa = 4.34WGPEE955 pKa = 2.86ICRR958 pKa = 11.84RR959 pKa = 11.84MRR961 pKa = 11.84SAYY964 pKa = 8.71WMFDD968 pKa = 3.15KK969 pKa = 10.48EE970 pKa = 4.7HH971 pKa = 7.14PPMAWGRR978 pKa = 11.84GISAALLVAGRR989 pKa = 11.84HH990 pKa = 4.53VPVLRR995 pKa = 11.84DD996 pKa = 2.77ICTWHH1001 pKa = 6.98LSVTHH1006 pKa = 6.07GPIMKK1011 pKa = 10.36LPFTNIWSTFYY1022 pKa = 10.32RR1023 pKa = 11.84YY1024 pKa = 9.01EE1025 pKa = 3.91VSGEE1029 pKa = 3.89FTDD1032 pKa = 4.31RR1033 pKa = 11.84GLNEE1037 pKa = 3.96FLADD1041 pKa = 3.8YY1042 pKa = 10.36DD1043 pKa = 3.82ISTSEE1048 pKa = 4.0YY1049 pKa = 10.66LDD1051 pKa = 4.09FVDD1054 pKa = 5.38KK1055 pKa = 11.3LKK1057 pKa = 11.38GCMDD1061 pKa = 4.27PFINIQHH1068 pKa = 5.86SVIQKK1073 pKa = 10.15LLMRR1077 pKa = 11.84EE1078 pKa = 4.05
Molecular weight: 122.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL64|A0A1L3KL64_9VIRU RNA-dependent RNA polymerase OS=Wenzhou yanvirus-like virus 1 OX=1923683 PE=4 SV=1
MM1 pKa = 6.33QQKK4 pKa = 9.0TSILPSTHH12 pKa = 6.65PVRR15 pKa = 11.84SLAGLARR22 pKa = 11.84QIALPHH28 pKa = 6.06EE29 pKa = 4.76FAPEE33 pKa = 4.18RR34 pKa = 11.84FPSFPALEE42 pKa = 4.15RR43 pKa = 11.84TAVMGFNVPSTWDD56 pKa = 3.48LPAASTVKK64 pKa = 10.78AILTRR69 pKa = 11.84QATFPFWFEE78 pKa = 3.94KK79 pKa = 9.28STSGATFRR87 pKa = 11.84VTWRR91 pKa = 11.84FPRR94 pKa = 11.84APGSNTTAYY103 pKa = 10.76DD104 pKa = 3.58PVINAAGTGNVAQTINQVGVTNGNNGGIASYY135 pKa = 9.68PILGLDD141 pKa = 4.25GMGTCPYY148 pKa = 10.09MYY150 pKa = 10.4IPDD153 pKa = 3.61NWTALMVVSLDD164 pKa = 3.62VAPATAIDD172 pKa = 3.69CTVIYY177 pKa = 9.77EE178 pKa = 4.43RR179 pKa = 11.84WNSPGQASVNSVVAGSTIAGNAGCSIPISMPSSEE213 pKa = 4.45STWIRR218 pKa = 11.84IVSVSLSTSTAIALGATNVTVVASSGTLAYY248 pKa = 9.42TPGVLAGNVQVTPAATVALLPGAYY272 pKa = 8.71PVEE275 pKa = 4.55FANSQLPWYY284 pKa = 7.41STRR287 pKa = 11.84TTAAAVLCTNVSQVLNKK304 pKa = 10.5GGTVLAGRR312 pKa = 11.84IAPQVVNPWNVSEE325 pKa = 4.3TYY327 pKa = 10.65INGLHH332 pKa = 6.3PAEE335 pKa = 4.6KK336 pKa = 10.49AFLPLEE342 pKa = 4.39TGLYY346 pKa = 9.76SYY348 pKa = 10.26CPPSTDD354 pKa = 5.79LVDD357 pKa = 4.2FWDD360 pKa = 3.8YY361 pKa = 10.69TLNSTPFFTLPATPLYY377 pKa = 10.68RR378 pKa = 11.84LDD380 pKa = 3.46NTSLVNVMFVTASAVAEE397 pKa = 4.11TLAVNVDD404 pKa = 2.69WHH406 pKa = 7.65IEE408 pKa = 3.85FRR410 pKa = 11.84TSSSLFPIGLSAVTLEE426 pKa = 4.51TLHH429 pKa = 5.81QAQLALVSAGFFFEE443 pKa = 4.42NPEE446 pKa = 4.16HH447 pKa = 6.84KK448 pKa = 10.32SVLSKK453 pKa = 11.45VMTAVKK459 pKa = 10.12RR460 pKa = 11.84YY461 pKa = 9.92GPTALGAVNPAAGRR475 pKa = 11.84VARR478 pKa = 11.84TMIQLSSKK486 pKa = 9.2PASRR490 pKa = 11.84MKK492 pKa = 9.27PTSASASGFNGNKK505 pKa = 9.6KK506 pKa = 9.89PPRR509 pKa = 11.84PTPKK513 pKa = 9.87KK514 pKa = 10.61GKK516 pKa = 8.61ATSGKK521 pKa = 9.64KK522 pKa = 9.85KK523 pKa = 10.29KK524 pKa = 10.41
MM1 pKa = 6.33QQKK4 pKa = 9.0TSILPSTHH12 pKa = 6.65PVRR15 pKa = 11.84SLAGLARR22 pKa = 11.84QIALPHH28 pKa = 6.06EE29 pKa = 4.76FAPEE33 pKa = 4.18RR34 pKa = 11.84FPSFPALEE42 pKa = 4.15RR43 pKa = 11.84TAVMGFNVPSTWDD56 pKa = 3.48LPAASTVKK64 pKa = 10.78AILTRR69 pKa = 11.84QATFPFWFEE78 pKa = 3.94KK79 pKa = 9.28STSGATFRR87 pKa = 11.84VTWRR91 pKa = 11.84FPRR94 pKa = 11.84APGSNTTAYY103 pKa = 10.76DD104 pKa = 3.58PVINAAGTGNVAQTINQVGVTNGNNGGIASYY135 pKa = 9.68PILGLDD141 pKa = 4.25GMGTCPYY148 pKa = 10.09MYY150 pKa = 10.4IPDD153 pKa = 3.61NWTALMVVSLDD164 pKa = 3.62VAPATAIDD172 pKa = 3.69CTVIYY177 pKa = 9.77EE178 pKa = 4.43RR179 pKa = 11.84WNSPGQASVNSVVAGSTIAGNAGCSIPISMPSSEE213 pKa = 4.45STWIRR218 pKa = 11.84IVSVSLSTSTAIALGATNVTVVASSGTLAYY248 pKa = 9.42TPGVLAGNVQVTPAATVALLPGAYY272 pKa = 8.71PVEE275 pKa = 4.55FANSQLPWYY284 pKa = 7.41STRR287 pKa = 11.84TTAAAVLCTNVSQVLNKK304 pKa = 10.5GGTVLAGRR312 pKa = 11.84IAPQVVNPWNVSEE325 pKa = 4.3TYY327 pKa = 10.65INGLHH332 pKa = 6.3PAEE335 pKa = 4.6KK336 pKa = 10.49AFLPLEE342 pKa = 4.39TGLYY346 pKa = 9.76SYY348 pKa = 10.26CPPSTDD354 pKa = 5.79LVDD357 pKa = 4.2FWDD360 pKa = 3.8YY361 pKa = 10.69TLNSTPFFTLPATPLYY377 pKa = 10.68RR378 pKa = 11.84LDD380 pKa = 3.46NTSLVNVMFVTASAVAEE397 pKa = 4.11TLAVNVDD404 pKa = 2.69WHH406 pKa = 7.65IEE408 pKa = 3.85FRR410 pKa = 11.84TSSSLFPIGLSAVTLEE426 pKa = 4.51TLHH429 pKa = 5.81QAQLALVSAGFFFEE443 pKa = 4.42NPEE446 pKa = 4.16HH447 pKa = 6.84KK448 pKa = 10.32SVLSKK453 pKa = 11.45VMTAVKK459 pKa = 10.12RR460 pKa = 11.84YY461 pKa = 9.92GPTALGAVNPAAGRR475 pKa = 11.84VARR478 pKa = 11.84TMIQLSSKK486 pKa = 9.2PASRR490 pKa = 11.84MKK492 pKa = 9.27PTSASASGFNGNKK505 pKa = 9.6KK506 pKa = 9.89PPRR509 pKa = 11.84PTPKK513 pKa = 9.87KK514 pKa = 10.61GKK516 pKa = 8.61ATSGKK521 pKa = 9.64KK522 pKa = 9.85KK523 pKa = 10.29KK524 pKa = 10.41
Molecular weight: 55.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1602 |
524 |
1078 |
801.0 |
89.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.176 ± 1.481 | 2.31 ± 0.756 |
3.62 ± 0.848 | 3.87 ± 0.562 |
4.744 ± 0.411 | 6.492 ± 0.211 |
2.31 ± 0.649 | 5.056 ± 0.585 |
3.371 ± 0.142 | 7.553 ± 0.151 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.559 ± 0.363 | 4.057 ± 0.717 |
6.866 ± 0.747 | 2.871 ± 0.111 |
6.117 ± 1.389 | 8.052 ± 0.724 |
6.617 ± 1.95 | 7.615 ± 0.755 |
2.185 ± 0.154 | 4.557 ± 1.051 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
