
Streptococcus satellite phage Javan341
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
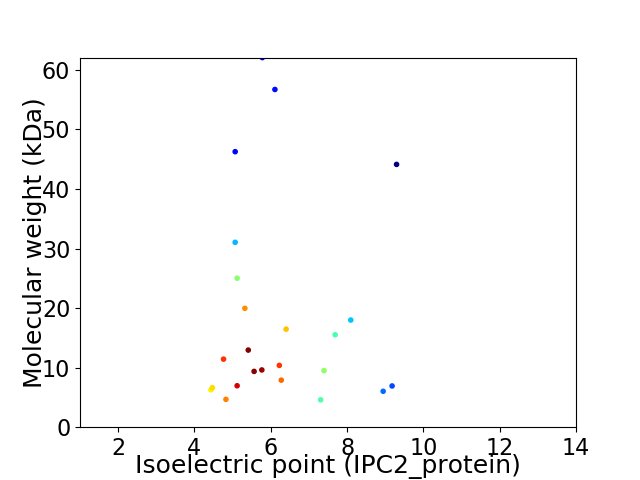
Virtual 2D-PAGE plot for 24 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
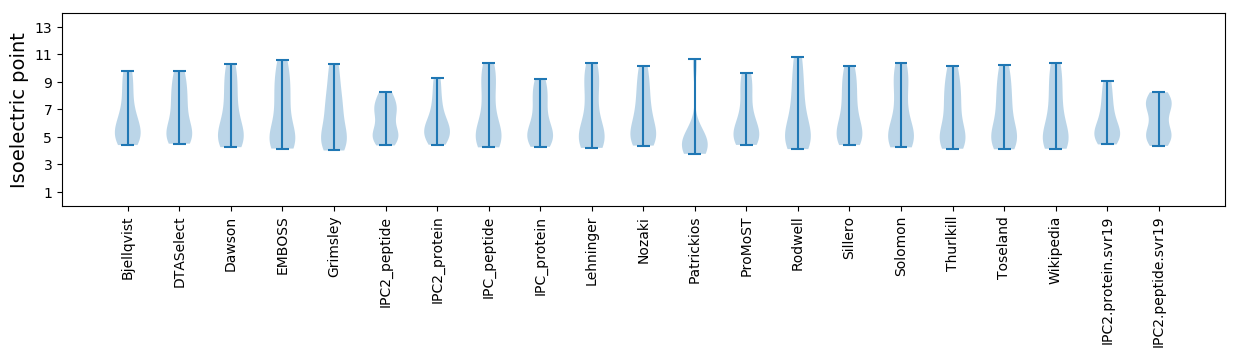
Protein with the lowest isoelectric point:
>tr|A0A4D5ZKZ1|A0A4D5ZKZ1_9VIRU HTH cro/C1-type domain-containing protein OS=Streptococcus satellite phage Javan341 OX=2558649 GN=JavanS341_0021 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 10.3IKK4 pKa = 10.74LFYY7 pKa = 10.44QKK9 pKa = 10.61YY10 pKa = 8.31KK11 pKa = 10.71QSLEE15 pKa = 4.1DD16 pKa = 4.85FEE18 pKa = 5.91SQVNDD23 pKa = 3.44FMATVEE29 pKa = 4.3VVDD32 pKa = 3.98VKK34 pKa = 11.25YY35 pKa = 11.23SEE37 pKa = 4.27ATVGNSDD44 pKa = 5.52DD45 pKa = 4.23MDD47 pKa = 4.02TLTSVMILYY56 pKa = 9.79KK57 pKa = 10.73
MM1 pKa = 7.66KK2 pKa = 10.3IKK4 pKa = 10.74LFYY7 pKa = 10.44QKK9 pKa = 10.61YY10 pKa = 8.31KK11 pKa = 10.71QSLEE15 pKa = 4.1DD16 pKa = 4.85FEE18 pKa = 5.91SQVNDD23 pKa = 3.44FMATVEE29 pKa = 4.3VVDD32 pKa = 3.98VKK34 pKa = 11.25YY35 pKa = 11.23SEE37 pKa = 4.27ATVGNSDD44 pKa = 5.52DD45 pKa = 4.23MDD47 pKa = 4.02TLTSVMILYY56 pKa = 9.79KK57 pKa = 10.73
Molecular weight: 6.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZQG3|A0A4D5ZQG3_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan341 OX=2558649 GN=JavanS341_0010 PE=4 SV=1
MM1 pKa = 7.92KK2 pKa = 9.25ITEE5 pKa = 4.18YY6 pKa = 10.59KK7 pKa = 9.86KK8 pKa = 10.3KK9 pKa = 10.54NGTIVYY15 pKa = 8.89RR16 pKa = 11.84ASVYY20 pKa = 10.88LGVDD24 pKa = 2.75KK25 pKa = 10.61LTGKK29 pKa = 10.25KK30 pKa = 10.31ARR32 pKa = 11.84TTVTAKK38 pKa = 9.59TKK40 pKa = 10.08TGIKK44 pKa = 9.52IKK46 pKa = 10.63ARR48 pKa = 11.84EE49 pKa = 4.43AINAFANNGYY59 pKa = 8.06SVKK62 pKa = 10.07EE63 pKa = 4.01KK64 pKa = 9.58PTITTYY70 pKa = 11.1KK71 pKa = 10.28EE72 pKa = 4.7LVALWWEE79 pKa = 4.57SYY81 pKa = 11.28KK82 pKa = 10.03NTIKK86 pKa = 10.78PNSQQSMEE94 pKa = 4.61GIVRR98 pKa = 11.84LHH100 pKa = 6.54ILPVFGDD107 pKa = 3.7YY108 pKa = 11.03KK109 pKa = 10.72LDD111 pKa = 3.64KK112 pKa = 9.61LTTPIIQQQVNKK124 pKa = 9.58WADD127 pKa = 3.33KK128 pKa = 10.63ANKK131 pKa = 9.16GEE133 pKa = 3.98KK134 pKa = 9.51GAYY137 pKa = 9.7ANYY140 pKa = 10.84SFLNNINRR148 pKa = 11.84RR149 pKa = 11.84ILQYY153 pKa = 10.72GVTMQAIQHH162 pKa = 5.58NPARR166 pKa = 11.84DD167 pKa = 3.51VIIPRR172 pKa = 11.84KK173 pKa = 7.62QQNKK177 pKa = 5.06EE178 pKa = 3.84HH179 pKa = 6.35KK180 pKa = 10.3VKK182 pKa = 10.6FFSNQEE188 pKa = 3.76LKK190 pKa = 10.87QFLDD194 pKa = 3.59YY195 pKa = 11.26LDD197 pKa = 5.03NLDD200 pKa = 3.77LSSYY204 pKa = 10.93EE205 pKa = 3.9NLFDD209 pKa = 3.71YY210 pKa = 10.96VLYY213 pKa = 8.9KK214 pKa = 10.39TLLASGCRR222 pKa = 11.84IGEE225 pKa = 4.0ALALEE230 pKa = 4.49WSDD233 pKa = 3.5IDD235 pKa = 4.04LKK237 pKa = 11.17KK238 pKa = 11.01GIISISKK245 pKa = 7.96TLNRR249 pKa = 11.84YY250 pKa = 8.7QEE252 pKa = 4.35TNTPKK257 pKa = 10.57SKK259 pKa = 10.86AGLRR263 pKa = 11.84EE264 pKa = 3.44IDD266 pKa = 3.3IDD268 pKa = 3.65KK269 pKa = 9.94ATISLLKK276 pKa = 10.08QYY278 pKa = 10.82KK279 pKa = 9.6KK280 pKa = 10.25RR281 pKa = 11.84QQVQSWQLGRR291 pKa = 11.84SEE293 pKa = 5.54GIVFTPFTTKK303 pKa = 10.34YY304 pKa = 10.75AYY306 pKa = 10.29ACLLRR311 pKa = 11.84KK312 pKa = 9.65RR313 pKa = 11.84LQGHH317 pKa = 6.88LKK319 pKa = 10.21SAGVPDD325 pKa = 3.89ISFHH329 pKa = 5.86GFRR332 pKa = 11.84HH333 pKa = 4.4THH335 pKa = 4.65ATIMLYY341 pKa = 10.62AGIEE345 pKa = 4.18AKK347 pKa = 10.42DD348 pKa = 3.35LQYY351 pKa = 11.55RR352 pKa = 11.84LGHH355 pKa = 6.02SNISMTLNTYY365 pKa = 8.7VHH367 pKa = 6.14ATKK370 pKa = 10.47EE371 pKa = 4.25GAKK374 pKa = 9.7KK375 pKa = 10.13AVSIFEE381 pKa = 4.24SAISNLL387 pKa = 3.62
MM1 pKa = 7.92KK2 pKa = 9.25ITEE5 pKa = 4.18YY6 pKa = 10.59KK7 pKa = 9.86KK8 pKa = 10.3KK9 pKa = 10.54NGTIVYY15 pKa = 8.89RR16 pKa = 11.84ASVYY20 pKa = 10.88LGVDD24 pKa = 2.75KK25 pKa = 10.61LTGKK29 pKa = 10.25KK30 pKa = 10.31ARR32 pKa = 11.84TTVTAKK38 pKa = 9.59TKK40 pKa = 10.08TGIKK44 pKa = 9.52IKK46 pKa = 10.63ARR48 pKa = 11.84EE49 pKa = 4.43AINAFANNGYY59 pKa = 8.06SVKK62 pKa = 10.07EE63 pKa = 4.01KK64 pKa = 9.58PTITTYY70 pKa = 11.1KK71 pKa = 10.28EE72 pKa = 4.7LVALWWEE79 pKa = 4.57SYY81 pKa = 11.28KK82 pKa = 10.03NTIKK86 pKa = 10.78PNSQQSMEE94 pKa = 4.61GIVRR98 pKa = 11.84LHH100 pKa = 6.54ILPVFGDD107 pKa = 3.7YY108 pKa = 11.03KK109 pKa = 10.72LDD111 pKa = 3.64KK112 pKa = 9.61LTTPIIQQQVNKK124 pKa = 9.58WADD127 pKa = 3.33KK128 pKa = 10.63ANKK131 pKa = 9.16GEE133 pKa = 3.98KK134 pKa = 9.51GAYY137 pKa = 9.7ANYY140 pKa = 10.84SFLNNINRR148 pKa = 11.84RR149 pKa = 11.84ILQYY153 pKa = 10.72GVTMQAIQHH162 pKa = 5.58NPARR166 pKa = 11.84DD167 pKa = 3.51VIIPRR172 pKa = 11.84KK173 pKa = 7.62QQNKK177 pKa = 5.06EE178 pKa = 3.84HH179 pKa = 6.35KK180 pKa = 10.3VKK182 pKa = 10.6FFSNQEE188 pKa = 3.76LKK190 pKa = 10.87QFLDD194 pKa = 3.59YY195 pKa = 11.26LDD197 pKa = 5.03NLDD200 pKa = 3.77LSSYY204 pKa = 10.93EE205 pKa = 3.9NLFDD209 pKa = 3.71YY210 pKa = 10.96VLYY213 pKa = 8.9KK214 pKa = 10.39TLLASGCRR222 pKa = 11.84IGEE225 pKa = 4.0ALALEE230 pKa = 4.49WSDD233 pKa = 3.5IDD235 pKa = 4.04LKK237 pKa = 11.17KK238 pKa = 11.01GIISISKK245 pKa = 7.96TLNRR249 pKa = 11.84YY250 pKa = 8.7QEE252 pKa = 4.35TNTPKK257 pKa = 10.57SKK259 pKa = 10.86AGLRR263 pKa = 11.84EE264 pKa = 3.44IDD266 pKa = 3.3IDD268 pKa = 3.65KK269 pKa = 9.94ATISLLKK276 pKa = 10.08QYY278 pKa = 10.82KK279 pKa = 9.6KK280 pKa = 10.25RR281 pKa = 11.84QQVQSWQLGRR291 pKa = 11.84SEE293 pKa = 5.54GIVFTPFTTKK303 pKa = 10.34YY304 pKa = 10.75AYY306 pKa = 10.29ACLLRR311 pKa = 11.84KK312 pKa = 9.65RR313 pKa = 11.84LQGHH317 pKa = 6.88LKK319 pKa = 10.21SAGVPDD325 pKa = 3.89ISFHH329 pKa = 5.86GFRR332 pKa = 11.84HH333 pKa = 4.4THH335 pKa = 4.65ATIMLYY341 pKa = 10.62AGIEE345 pKa = 4.18AKK347 pKa = 10.42DD348 pKa = 3.35LQYY351 pKa = 11.55RR352 pKa = 11.84LGHH355 pKa = 6.02SNISMTLNTYY365 pKa = 8.7VHH367 pKa = 6.14ATKK370 pKa = 10.47EE371 pKa = 4.25GAKK374 pKa = 9.7KK375 pKa = 10.13AVSIFEE381 pKa = 4.24SAISNLL387 pKa = 3.62
Molecular weight: 44.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3884 |
39 |
536 |
161.8 |
18.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.334 ± 0.608 | 0.335 ± 0.085 |
6.54 ± 0.565 | 8.883 ± 0.79 |
4.222 ± 0.437 | 5.046 ± 0.491 |
1.287 ± 0.201 | 7.055 ± 0.611 |
9.604 ± 0.469 | 10.067 ± 0.557 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.214 ± 0.32 | 5.252 ± 0.346 |
2.111 ± 0.355 | 4.737 ± 0.687 |
5.227 ± 0.339 | 5.664 ± 0.401 |
4.866 ± 0.509 | 5.407 ± 0.585 |
0.747 ± 0.139 | 4.403 ± 0.381 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
