
Cucumber mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
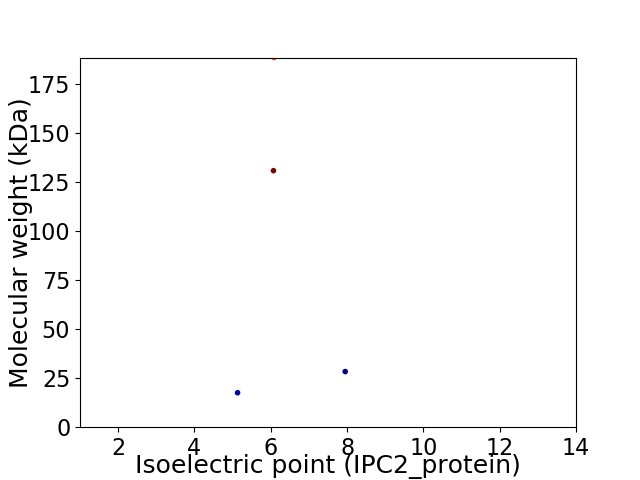
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0PA52|A0PA52_9VIRU Isoform of A0PA51 130 kDa replicase OS=Cucumber mottle virus OX=388038 GN=replicase PE=4 SV=1
MM1 pKa = 7.9AYY3 pKa = 10.47SPITPTNLSLFSSNYY18 pKa = 8.82VPFTEE23 pKa = 5.35FYY25 pKa = 10.18NYY27 pKa = 10.6LVTAQGEE34 pKa = 4.36AFQTQQGRR42 pKa = 11.84DD43 pKa = 3.42SVRR46 pKa = 11.84DD47 pKa = 3.6SLSGFFSSPVSPTVRR62 pKa = 11.84FPDD65 pKa = 3.56GVFYY69 pKa = 11.23VFLGNPVLDD78 pKa = 4.34PLFKK82 pKa = 10.9ALLQSLDD89 pKa = 3.02TRR91 pKa = 11.84NRR93 pKa = 11.84VIEE96 pKa = 4.0VDD98 pKa = 3.49NPSNPTTAEE107 pKa = 3.83SLNAVQRR114 pKa = 11.84TDD116 pKa = 3.3DD117 pKa = 3.72STVSSRR123 pKa = 11.84VGLINLRR130 pKa = 11.84AAITQGNGVVNRR142 pKa = 11.84SIFEE146 pKa = 4.15SSNGLTWATSGSSSSKK162 pKa = 10.73
MM1 pKa = 7.9AYY3 pKa = 10.47SPITPTNLSLFSSNYY18 pKa = 8.82VPFTEE23 pKa = 5.35FYY25 pKa = 10.18NYY27 pKa = 10.6LVTAQGEE34 pKa = 4.36AFQTQQGRR42 pKa = 11.84DD43 pKa = 3.42SVRR46 pKa = 11.84DD47 pKa = 3.6SLSGFFSSPVSPTVRR62 pKa = 11.84FPDD65 pKa = 3.56GVFYY69 pKa = 11.23VFLGNPVLDD78 pKa = 4.34PLFKK82 pKa = 10.9ALLQSLDD89 pKa = 3.02TRR91 pKa = 11.84NRR93 pKa = 11.84VIEE96 pKa = 4.0VDD98 pKa = 3.49NPSNPTTAEE107 pKa = 3.83SLNAVQRR114 pKa = 11.84TDD116 pKa = 3.3DD117 pKa = 3.72STVSSRR123 pKa = 11.84VGLINLRR130 pKa = 11.84AAITQGNGVVNRR142 pKa = 11.84SIFEE146 pKa = 4.15SSNGLTWATSGSSSSKK162 pKa = 10.73
Molecular weight: 17.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0PA54|A0PA54_9VIRU Capsid protein OS=Cucumber mottle virus OX=388038 GN=cp PE=3 SV=1
MM1 pKa = 7.58SISNLSVEE9 pKa = 4.59SQLKK13 pKa = 9.49PSNFVKK19 pKa = 10.73LSWVDD24 pKa = 3.76KK25 pKa = 10.66LLPDD29 pKa = 3.55YY30 pKa = 10.88FSILRR35 pKa = 11.84FLSVTDD41 pKa = 3.5HH42 pKa = 6.36SVIKK46 pKa = 10.49ARR48 pKa = 11.84EE49 pKa = 4.03YY50 pKa = 10.88EE51 pKa = 4.1SFLPVEE57 pKa = 4.47LLRR60 pKa = 11.84GVDD63 pKa = 3.87LTKK66 pKa = 10.6HH67 pKa = 6.03KK68 pKa = 10.85YY69 pKa = 7.96VTLLGVVISGVWTIPEE85 pKa = 4.58GCSGGATVGLVDD97 pKa = 3.4TRR99 pKa = 11.84MEE101 pKa = 4.01RR102 pKa = 11.84VVEE105 pKa = 4.04GTVCKK110 pKa = 10.39FSVPASVRR118 pKa = 11.84EE119 pKa = 3.9FNVRR123 pKa = 11.84FIPNYY128 pKa = 10.46SITAADD134 pKa = 4.09AARR137 pKa = 11.84HH138 pKa = 5.09PWSLFVRR145 pKa = 11.84LKK147 pKa = 10.62GVNIKK152 pKa = 10.87DD153 pKa = 3.64SFSPLTLEE161 pKa = 4.01IAALVATTNSIIKK174 pKa = 9.87KK175 pKa = 7.36SLKK178 pKa = 10.51AIVSDD183 pKa = 3.85VVVGSDD189 pKa = 3.19AAVAIADD196 pKa = 4.42RR197 pKa = 11.84DD198 pKa = 3.95SQVNSFFDD206 pKa = 3.57SVPITKK212 pKa = 10.2SVVNFDD218 pKa = 3.23KK219 pKa = 10.7SYY221 pKa = 10.83KK222 pKa = 10.24SRR224 pKa = 11.84VPKK227 pKa = 10.05KK228 pKa = 10.19DD229 pKa = 3.16SSGGSGKK236 pKa = 9.75VKK238 pKa = 10.5RR239 pKa = 11.84STGTSGPADD248 pKa = 3.57TEE250 pKa = 4.21FSDD253 pKa = 6.06DD254 pKa = 3.81GLLSNHH260 pKa = 6.27SDD262 pKa = 3.01
MM1 pKa = 7.58SISNLSVEE9 pKa = 4.59SQLKK13 pKa = 9.49PSNFVKK19 pKa = 10.73LSWVDD24 pKa = 3.76KK25 pKa = 10.66LLPDD29 pKa = 3.55YY30 pKa = 10.88FSILRR35 pKa = 11.84FLSVTDD41 pKa = 3.5HH42 pKa = 6.36SVIKK46 pKa = 10.49ARR48 pKa = 11.84EE49 pKa = 4.03YY50 pKa = 10.88EE51 pKa = 4.1SFLPVEE57 pKa = 4.47LLRR60 pKa = 11.84GVDD63 pKa = 3.87LTKK66 pKa = 10.6HH67 pKa = 6.03KK68 pKa = 10.85YY69 pKa = 7.96VTLLGVVISGVWTIPEE85 pKa = 4.58GCSGGATVGLVDD97 pKa = 3.4TRR99 pKa = 11.84MEE101 pKa = 4.01RR102 pKa = 11.84VVEE105 pKa = 4.04GTVCKK110 pKa = 10.39FSVPASVRR118 pKa = 11.84EE119 pKa = 3.9FNVRR123 pKa = 11.84FIPNYY128 pKa = 10.46SITAADD134 pKa = 4.09AARR137 pKa = 11.84HH138 pKa = 5.09PWSLFVRR145 pKa = 11.84LKK147 pKa = 10.62GVNIKK152 pKa = 10.87DD153 pKa = 3.64SFSPLTLEE161 pKa = 4.01IAALVATTNSIIKK174 pKa = 9.87KK175 pKa = 7.36SLKK178 pKa = 10.51AIVSDD183 pKa = 3.85VVVGSDD189 pKa = 3.19AAVAIADD196 pKa = 4.42RR197 pKa = 11.84DD198 pKa = 3.95SQVNSFFDD206 pKa = 3.57SVPITKK212 pKa = 10.2SVVNFDD218 pKa = 3.23KK219 pKa = 10.7SYY221 pKa = 10.83KK222 pKa = 10.24SRR224 pKa = 11.84VPKK227 pKa = 10.05KK228 pKa = 10.19DD229 pKa = 3.16SSGGSGKK236 pKa = 9.75VKK238 pKa = 10.5RR239 pKa = 11.84STGTSGPADD248 pKa = 3.57TEE250 pKa = 4.21FSDD253 pKa = 6.06DD254 pKa = 3.81GLLSNHH260 pKa = 6.27SDD262 pKa = 3.01
Molecular weight: 28.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3262 |
162 |
1672 |
815.5 |
91.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.499 ± 0.256 | 1.594 ± 0.429 |
6.683 ± 0.324 | 4.997 ± 0.569 |
6.101 ± 0.26 | 4.844 ± 0.469 |
2.115 ± 0.482 | 4.966 ± 0.328 |
6.407 ± 0.91 | 8.768 ± 0.303 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.931 ± 0.447 | 4.69 ± 0.461 |
4.384 ± 0.322 | 2.606 ± 0.417 |
4.782 ± 0.173 | 8.829 ± 1.956 |
5.917 ± 0.504 | 9.718 ± 0.698 |
0.95 ± 0.059 | 3.188 ± 0.308 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
