
Nitratireductor indicus C115
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Nitratireductor; Nitratireductor indicus
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
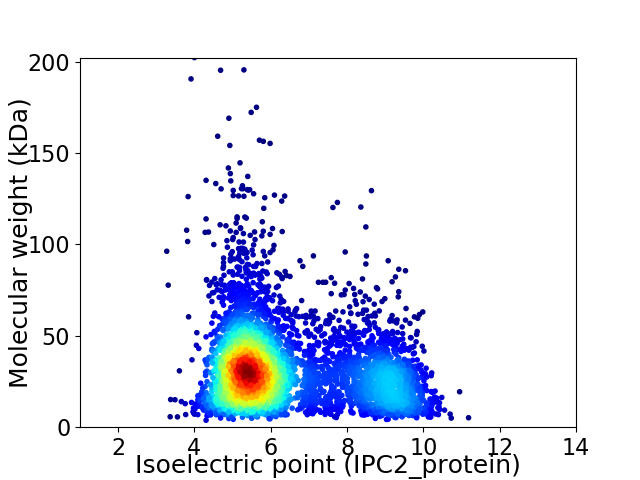
Virtual 2D-PAGE plot for 4821 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K2NXR5|K2NXR5_9RHIZ Uncharacterized protein OS=Nitratireductor indicus C115 OX=1231190 GN=NA8A_04365 PE=4 SV=1
MM1 pKa = 7.73EE2 pKa = 4.42VRR4 pKa = 11.84QSSGAGIVDD13 pKa = 3.4WNGFSIGSDD22 pKa = 3.09NQVHH26 pKa = 6.2FEE28 pKa = 4.13NGSGATLNRR37 pKa = 11.84VTGNVSSQIDD47 pKa = 3.82GALTATGSVFLINPAGVVVGSGGMVATGGSFVASTHH83 pKa = 6.67DD84 pKa = 3.4VTDD87 pKa = 3.72ADD89 pKa = 4.16FLDD92 pKa = 4.49GGSLIFSGTSEE103 pKa = 4.45AEE105 pKa = 4.06VVNAGTIRR113 pKa = 11.84SQQGDD118 pKa = 3.14IALIARR124 pKa = 11.84RR125 pKa = 11.84VEE127 pKa = 3.89NSGTLEE133 pKa = 4.31APNGTAGLAAGYY145 pKa = 9.31KK146 pKa = 10.25VLMKK150 pKa = 10.68DD151 pKa = 3.07STDD154 pKa = 3.59ADD156 pKa = 3.82GLLSVQLGGEE166 pKa = 4.06DD167 pKa = 3.54TEE169 pKa = 4.72AVNSGTIAAANAEE182 pKa = 3.72IRR184 pKa = 11.84ANGGNVYY191 pKa = 10.71ALAGNTEE198 pKa = 4.37GVIKK202 pKa = 9.99ATGVTSSGGRR212 pKa = 11.84IFLTAGAAGKK222 pKa = 10.56VKK224 pKa = 9.67VTGRR228 pKa = 11.84LKK230 pKa = 11.01ARR232 pKa = 11.84TLPKK236 pKa = 9.2TAPVPTPRR244 pKa = 11.84PATEE248 pKa = 4.57GGHH251 pKa = 4.44VTVSGGDD258 pKa = 3.24ISISGTIDD266 pKa = 2.99ASAEE270 pKa = 4.01TSGTKK275 pKa = 10.34GGTVTAIASSGMSFSGAIEE294 pKa = 4.12AKK296 pKa = 10.26GGAGGSGGFVEE307 pKa = 5.03TSGEE311 pKa = 4.31SISIADD317 pKa = 3.92TARR320 pKa = 11.84VTTLSGGGEE329 pKa = 4.09AGTWLIDD336 pKa = 4.13PNDD339 pKa = 3.68LTIAASGGDD348 pKa = 3.3ITGEE352 pKa = 4.04TLSTNLAGGNVTLSSNDD369 pKa = 3.1GATDD373 pKa = 4.02GSGDD377 pKa = 3.34IFVNDD382 pKa = 5.11AISWTADD389 pKa = 2.91TTLTLNAVRR398 pKa = 11.84NVEE401 pKa = 4.05INDD404 pKa = 4.15TITATGTNAGLALTYY419 pKa = 10.78GGGYY423 pKa = 10.0SFANGGRR430 pKa = 11.84ITLSGDD436 pKa = 3.5SANLSLNGNAYY447 pKa = 10.57ALIHH451 pKa = 6.8DD452 pKa = 4.54VNDD455 pKa = 3.52LQAISGSGLYY465 pKa = 10.58ALAEE469 pKa = 4.52TIDD472 pKa = 3.77ASGTTSWNSGAGFNPISGFSGTLAGLGNTIEE503 pKa = 4.37SLTINRR509 pKa = 11.84PGEE512 pKa = 3.97ATVGMFSSTSGGFRR526 pKa = 11.84DD527 pKa = 4.17FTLSNVSIVSGGSVGALAYY546 pKa = 10.22RR547 pKa = 11.84FNAGQGASNIHH558 pKa = 4.5VTGTITSLGGGGQVGGLVGWSDD580 pKa = 3.23QTAISNSSSTATVTGFGEE598 pKa = 4.22VGGLVGRR605 pKa = 11.84LRR607 pKa = 11.84AASITNSYY615 pKa = 8.46ATGAVTGTTDD625 pKa = 3.37YY626 pKa = 11.62VGGLVGSTSQGGMLTNVYY644 pKa = 10.82ASGNVTGTTNVGGLVGGAILAGAVTATNAYY674 pKa = 8.65WDD676 pKa = 3.83VDD678 pKa = 3.74STGQSTSFAGTGIGNANAFTQSTYY702 pKa = 11.4SGFDD706 pKa = 3.34FTNTWVLIDD715 pKa = 3.73GEE717 pKa = 4.76TRR719 pKa = 11.84PMLQSEE725 pKa = 4.8YY726 pKa = 9.42STTIFTPHH734 pKa = 6.28QLQLMALDD742 pKa = 4.71LGADD746 pKa = 3.68YY747 pKa = 10.73MLGADD752 pKa = 4.91LDD754 pKa = 3.99MTGAFAANSGGYY766 pKa = 9.46YY767 pKa = 10.21GGVWSSAGFDD777 pKa = 3.33PVGINSANPFTGGLDD792 pKa = 3.97GQGHH796 pKa = 6.6TISGLKK802 pKa = 9.22IDD804 pKa = 5.07RR805 pKa = 11.84SGEE808 pKa = 4.11MNVGLLGYY816 pKa = 10.18ADD818 pKa = 4.01NATVSDD824 pKa = 4.04LTLSGGSIAGGQLVGALAGEE844 pKa = 4.52FNGGTITSVSSNISVAGSEE863 pKa = 4.38FVGGLVGASQNFTAISLASASGDD886 pKa = 3.36VSGSGDD892 pKa = 3.55TVGGLIGALFEE903 pKa = 4.53GTLSQSYY910 pKa = 8.07ATGNVSGSSLVGGLVGANGYY930 pKa = 10.76ASGGGTIDD938 pKa = 3.85QSFATGNVTGAGYY951 pKa = 11.09GLGGLVGSNEE961 pKa = 4.06GAISNAYY968 pKa = 10.48AMGNVTSTGASSATGGFVGVVLSSASIEE996 pKa = 3.95NAYY999 pKa = 9.07ATGKK1003 pKa = 8.29VTGAAPVGGFAGSSGADD1020 pKa = 2.98ASAIKK1025 pKa = 9.95SAYY1028 pKa = 8.63WNTEE1032 pKa = 3.36TSGQASGIVAGGGDD1046 pKa = 3.41ITGLTTAGLQGVLPSGFSNAVWGTGTDD1073 pKa = 3.91LYY1075 pKa = 10.51PYY1077 pKa = 9.92FNWLYY1082 pKa = 8.02PTGASAVSGFAYY1094 pKa = 10.57SDD1096 pKa = 3.4AGTTALTGAEE1106 pKa = 3.86IGIVSGGSFLGGTGTGANGYY1126 pKa = 9.38YY1127 pKa = 10.27YY1128 pKa = 11.17YY1129 pKa = 11.05LALPGALYY1137 pKa = 10.78ASGALAFLDD1146 pKa = 4.21GEE1148 pKa = 4.5AMQGAALGDD1157 pKa = 4.26SVTSTGVTGLDD1168 pKa = 3.13IYY1170 pKa = 11.19GSSLNLVTNEE1180 pKa = 3.91AALSDD1185 pKa = 3.8TVANLTTALGSYY1197 pKa = 9.21TDD1199 pKa = 3.97SDD1201 pKa = 4.23LAFIDD1206 pKa = 4.16TSATQLTASGQGLYY1220 pKa = 10.6LDD1222 pKa = 4.45AASDD1226 pKa = 3.79YY1227 pKa = 11.25TLDD1230 pKa = 3.58QDD1232 pKa = 4.08LTAGGLLSLTSGGTFTIDD1250 pKa = 3.69ADD1252 pKa = 4.24RR1253 pKa = 11.84TLKK1256 pKa = 8.03TTDD1259 pKa = 3.08AGSILVNSGVTWNGTGTLTLNAADD1283 pKa = 4.49NIALNGAII1291 pKa = 4.58
MM1 pKa = 7.73EE2 pKa = 4.42VRR4 pKa = 11.84QSSGAGIVDD13 pKa = 3.4WNGFSIGSDD22 pKa = 3.09NQVHH26 pKa = 6.2FEE28 pKa = 4.13NGSGATLNRR37 pKa = 11.84VTGNVSSQIDD47 pKa = 3.82GALTATGSVFLINPAGVVVGSGGMVATGGSFVASTHH83 pKa = 6.67DD84 pKa = 3.4VTDD87 pKa = 3.72ADD89 pKa = 4.16FLDD92 pKa = 4.49GGSLIFSGTSEE103 pKa = 4.45AEE105 pKa = 4.06VVNAGTIRR113 pKa = 11.84SQQGDD118 pKa = 3.14IALIARR124 pKa = 11.84RR125 pKa = 11.84VEE127 pKa = 3.89NSGTLEE133 pKa = 4.31APNGTAGLAAGYY145 pKa = 9.31KK146 pKa = 10.25VLMKK150 pKa = 10.68DD151 pKa = 3.07STDD154 pKa = 3.59ADD156 pKa = 3.82GLLSVQLGGEE166 pKa = 4.06DD167 pKa = 3.54TEE169 pKa = 4.72AVNSGTIAAANAEE182 pKa = 3.72IRR184 pKa = 11.84ANGGNVYY191 pKa = 10.71ALAGNTEE198 pKa = 4.37GVIKK202 pKa = 9.99ATGVTSSGGRR212 pKa = 11.84IFLTAGAAGKK222 pKa = 10.56VKK224 pKa = 9.67VTGRR228 pKa = 11.84LKK230 pKa = 11.01ARR232 pKa = 11.84TLPKK236 pKa = 9.2TAPVPTPRR244 pKa = 11.84PATEE248 pKa = 4.57GGHH251 pKa = 4.44VTVSGGDD258 pKa = 3.24ISISGTIDD266 pKa = 2.99ASAEE270 pKa = 4.01TSGTKK275 pKa = 10.34GGTVTAIASSGMSFSGAIEE294 pKa = 4.12AKK296 pKa = 10.26GGAGGSGGFVEE307 pKa = 5.03TSGEE311 pKa = 4.31SISIADD317 pKa = 3.92TARR320 pKa = 11.84VTTLSGGGEE329 pKa = 4.09AGTWLIDD336 pKa = 4.13PNDD339 pKa = 3.68LTIAASGGDD348 pKa = 3.3ITGEE352 pKa = 4.04TLSTNLAGGNVTLSSNDD369 pKa = 3.1GATDD373 pKa = 4.02GSGDD377 pKa = 3.34IFVNDD382 pKa = 5.11AISWTADD389 pKa = 2.91TTLTLNAVRR398 pKa = 11.84NVEE401 pKa = 4.05INDD404 pKa = 4.15TITATGTNAGLALTYY419 pKa = 10.78GGGYY423 pKa = 10.0SFANGGRR430 pKa = 11.84ITLSGDD436 pKa = 3.5SANLSLNGNAYY447 pKa = 10.57ALIHH451 pKa = 6.8DD452 pKa = 4.54VNDD455 pKa = 3.52LQAISGSGLYY465 pKa = 10.58ALAEE469 pKa = 4.52TIDD472 pKa = 3.77ASGTTSWNSGAGFNPISGFSGTLAGLGNTIEE503 pKa = 4.37SLTINRR509 pKa = 11.84PGEE512 pKa = 3.97ATVGMFSSTSGGFRR526 pKa = 11.84DD527 pKa = 4.17FTLSNVSIVSGGSVGALAYY546 pKa = 10.22RR547 pKa = 11.84FNAGQGASNIHH558 pKa = 4.5VTGTITSLGGGGQVGGLVGWSDD580 pKa = 3.23QTAISNSSSTATVTGFGEE598 pKa = 4.22VGGLVGRR605 pKa = 11.84LRR607 pKa = 11.84AASITNSYY615 pKa = 8.46ATGAVTGTTDD625 pKa = 3.37YY626 pKa = 11.62VGGLVGSTSQGGMLTNVYY644 pKa = 10.82ASGNVTGTTNVGGLVGGAILAGAVTATNAYY674 pKa = 8.65WDD676 pKa = 3.83VDD678 pKa = 3.74STGQSTSFAGTGIGNANAFTQSTYY702 pKa = 11.4SGFDD706 pKa = 3.34FTNTWVLIDD715 pKa = 3.73GEE717 pKa = 4.76TRR719 pKa = 11.84PMLQSEE725 pKa = 4.8YY726 pKa = 9.42STTIFTPHH734 pKa = 6.28QLQLMALDD742 pKa = 4.71LGADD746 pKa = 3.68YY747 pKa = 10.73MLGADD752 pKa = 4.91LDD754 pKa = 3.99MTGAFAANSGGYY766 pKa = 9.46YY767 pKa = 10.21GGVWSSAGFDD777 pKa = 3.33PVGINSANPFTGGLDD792 pKa = 3.97GQGHH796 pKa = 6.6TISGLKK802 pKa = 9.22IDD804 pKa = 5.07RR805 pKa = 11.84SGEE808 pKa = 4.11MNVGLLGYY816 pKa = 10.18ADD818 pKa = 4.01NATVSDD824 pKa = 4.04LTLSGGSIAGGQLVGALAGEE844 pKa = 4.52FNGGTITSVSSNISVAGSEE863 pKa = 4.38FVGGLVGASQNFTAISLASASGDD886 pKa = 3.36VSGSGDD892 pKa = 3.55TVGGLIGALFEE903 pKa = 4.53GTLSQSYY910 pKa = 8.07ATGNVSGSSLVGGLVGANGYY930 pKa = 10.76ASGGGTIDD938 pKa = 3.85QSFATGNVTGAGYY951 pKa = 11.09GLGGLVGSNEE961 pKa = 4.06GAISNAYY968 pKa = 10.48AMGNVTSTGASSATGGFVGVVLSSASIEE996 pKa = 3.95NAYY999 pKa = 9.07ATGKK1003 pKa = 8.29VTGAAPVGGFAGSSGADD1020 pKa = 2.98ASAIKK1025 pKa = 9.95SAYY1028 pKa = 8.63WNTEE1032 pKa = 3.36TSGQASGIVAGGGDD1046 pKa = 3.41ITGLTTAGLQGVLPSGFSNAVWGTGTDD1073 pKa = 3.91LYY1075 pKa = 10.51PYY1077 pKa = 9.92FNWLYY1082 pKa = 8.02PTGASAVSGFAYY1094 pKa = 10.57SDD1096 pKa = 3.4AGTTALTGAEE1106 pKa = 3.86IGIVSGGSFLGGTGTGANGYY1126 pKa = 9.38YY1127 pKa = 10.27YY1128 pKa = 11.17YY1129 pKa = 11.05LALPGALYY1137 pKa = 10.78ASGALAFLDD1146 pKa = 4.21GEE1148 pKa = 4.5AMQGAALGDD1157 pKa = 4.26SVTSTGVTGLDD1168 pKa = 3.13IYY1170 pKa = 11.19GSSLNLVTNEE1180 pKa = 3.91AALSDD1185 pKa = 3.8TVANLTTALGSYY1197 pKa = 9.21TDD1199 pKa = 3.97SDD1201 pKa = 4.23LAFIDD1206 pKa = 4.16TSATQLTASGQGLYY1220 pKa = 10.6LDD1222 pKa = 4.45AASDD1226 pKa = 3.79YY1227 pKa = 11.25TLDD1230 pKa = 3.58QDD1232 pKa = 4.08LTAGGLLSLTSGGTFTIDD1250 pKa = 3.69ADD1252 pKa = 4.24RR1253 pKa = 11.84TLKK1256 pKa = 8.03TTDD1259 pKa = 3.08AGSILVNSGVTWNGTGTLTLNAADD1283 pKa = 4.49NIALNGAII1291 pKa = 4.58
Molecular weight: 126.13 kDa
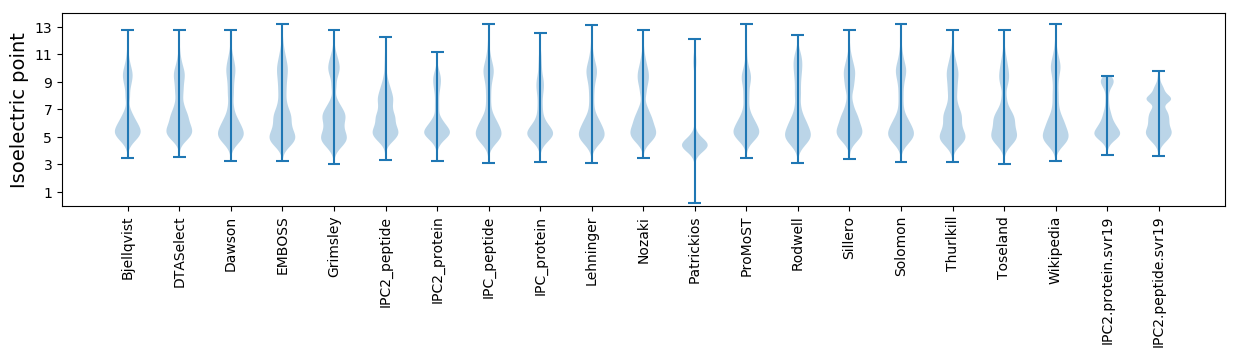
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K2NPJ2|K2NPJ2_9RHIZ Uncharacterized protein OS=Nitratireductor indicus C115 OX=1231190 GN=NA8A_24236 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.09QPSKK9 pKa = 8.8IVRR12 pKa = 11.84KK13 pKa = 9.24RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84RR29 pKa = 11.84VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.09QPSKK9 pKa = 8.8IVRR12 pKa = 11.84KK13 pKa = 9.24RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84RR29 pKa = 11.84VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1483311 |
32 |
1972 |
307.7 |
33.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.185 ± 0.043 | 0.823 ± 0.011 |
5.509 ± 0.032 | 6.182 ± 0.039 |
3.917 ± 0.023 | 8.674 ± 0.035 |
1.958 ± 0.018 | 5.455 ± 0.03 |
3.369 ± 0.029 | 10.075 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.651 ± 0.016 | 2.672 ± 0.017 |
4.942 ± 0.023 | 2.942 ± 0.021 |
7.07 ± 0.037 | 5.581 ± 0.027 |
5.152 ± 0.027 | 7.38 ± 0.027 |
1.257 ± 0.013 | 2.206 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
