
human papillomavirus 163
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 20
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
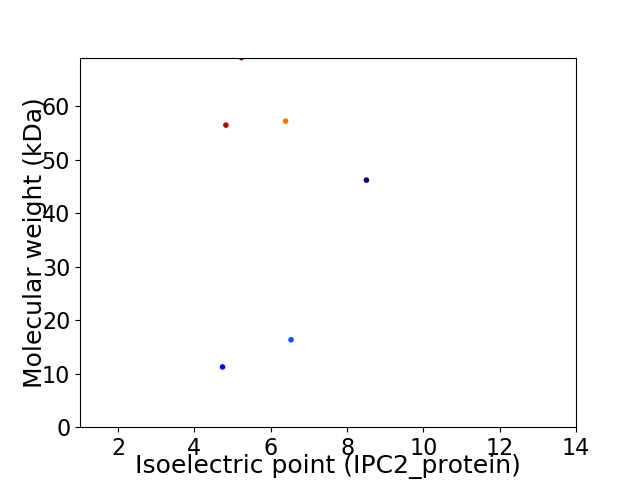
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
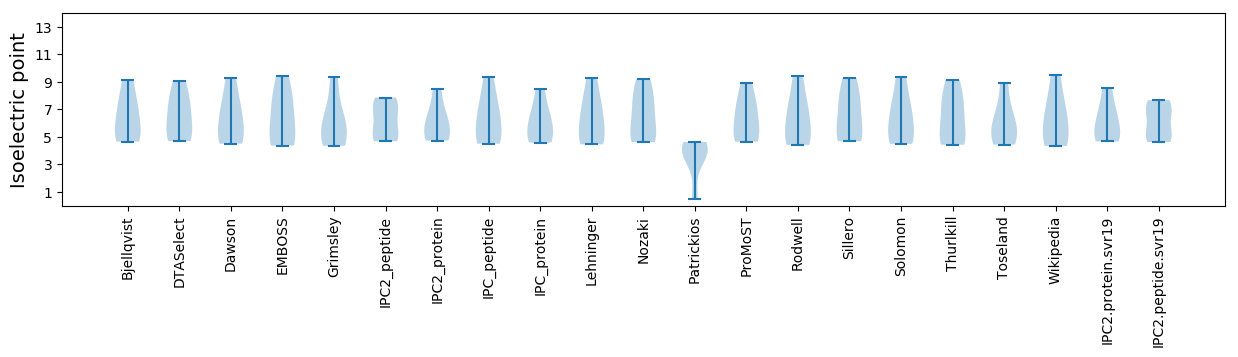
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4MYX6|K4MYX6_9PAPI Regulatory protein E2 OS=human papillomavirus 163 OX=1315262 GN=E2 PE=3 SV=1
MM1 pKa = 7.88RR2 pKa = 11.84GNAPNIQDD10 pKa = 3.16IEE12 pKa = 4.56LNLEE16 pKa = 4.09EE17 pKa = 5.32LVLPINVLSEE27 pKa = 4.14EE28 pKa = 4.64VIVSSDD34 pKa = 3.63DD35 pKa = 3.59EE36 pKa = 4.31EE37 pKa = 4.79QEE39 pKa = 3.94EE40 pKa = 4.84EE41 pKa = 4.24EE42 pKa = 4.26FVAYY46 pKa = 10.43RR47 pKa = 11.84IDD49 pKa = 3.93TCCHH53 pKa = 5.07SCRR56 pKa = 11.84ASVRR60 pKa = 11.84FTVFAKK66 pKa = 9.98TFAIRR71 pKa = 11.84LLEE74 pKa = 3.87QLILDD79 pKa = 4.48EE80 pKa = 5.62GLSLCCPSCARR91 pKa = 11.84NRR93 pKa = 11.84GRR95 pKa = 11.84NGRR98 pKa = 11.84RR99 pKa = 3.09
MM1 pKa = 7.88RR2 pKa = 11.84GNAPNIQDD10 pKa = 3.16IEE12 pKa = 4.56LNLEE16 pKa = 4.09EE17 pKa = 5.32LVLPINVLSEE27 pKa = 4.14EE28 pKa = 4.64VIVSSDD34 pKa = 3.63DD35 pKa = 3.59EE36 pKa = 4.31EE37 pKa = 4.79QEE39 pKa = 3.94EE40 pKa = 4.84EE41 pKa = 4.24EE42 pKa = 4.26FVAYY46 pKa = 10.43RR47 pKa = 11.84IDD49 pKa = 3.93TCCHH53 pKa = 5.07SCRR56 pKa = 11.84ASVRR60 pKa = 11.84FTVFAKK66 pKa = 9.98TFAIRR71 pKa = 11.84LLEE74 pKa = 3.87QLILDD79 pKa = 4.48EE80 pKa = 5.62GLSLCCPSCARR91 pKa = 11.84NRR93 pKa = 11.84GRR95 pKa = 11.84NGRR98 pKa = 11.84RR99 pKa = 3.09
Molecular weight: 11.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4MYX6|K4MYX6_9PAPI Regulatory protein E2 OS=human papillomavirus 163 OX=1315262 GN=E2 PE=3 SV=1
MM1 pKa = 8.34AMEE4 pKa = 4.65SLTDD8 pKa = 3.98RR9 pKa = 11.84FDD11 pKa = 3.95ALQEE15 pKa = 4.61LILKK19 pKa = 9.93HH20 pKa = 6.32IEE22 pKa = 3.92SDD24 pKa = 3.74STSLQSHH31 pKa = 5.16ITYY34 pKa = 8.85WGYY37 pKa = 10.2VRR39 pKa = 11.84KK40 pKa = 9.81EE41 pKa = 3.7NALMHH46 pKa = 6.2VARR49 pKa = 11.84QQGLTRR55 pKa = 11.84LGLQPLPALPVTEE68 pKa = 4.7YY69 pKa = 10.67NAKK72 pKa = 9.54QAITIQLTLQSLLRR86 pKa = 11.84SQYY89 pKa = 11.21GNEE92 pKa = 3.77PWTLTEE98 pKa = 4.67VSAEE102 pKa = 4.48LINTPPQNLLKK113 pKa = 10.34KK114 pKa = 10.28DD115 pKa = 3.72GFSVEE120 pKa = 3.77VWFDD124 pKa = 3.25NDD126 pKa = 3.08KK127 pKa = 11.13NNAMVYY133 pKa = 8.43TNWNDD138 pKa = 2.96MYY140 pKa = 11.26YY141 pKa = 10.69QDD143 pKa = 5.61AQEE146 pKa = 4.25TWHH149 pKa = 6.63KK150 pKa = 10.43VHH152 pKa = 7.14GSVDD156 pKa = 3.58YY157 pKa = 11.27NGLYY161 pKa = 8.8FTDD164 pKa = 3.91HH165 pKa = 6.19NGEE168 pKa = 3.88RR169 pKa = 11.84AYY171 pKa = 9.7FTLFDD176 pKa = 3.96TDD178 pKa = 4.61AIRR181 pKa = 11.84YY182 pKa = 6.27STTGLWTVHH191 pKa = 5.74YY192 pKa = 8.81KK193 pKa = 10.1SHH195 pKa = 6.57VISSSVVSSTTRR207 pKa = 11.84DD208 pKa = 3.15SSTTPEE214 pKa = 4.3DD215 pKa = 3.7SVDD218 pKa = 3.83RR219 pKa = 11.84PSTSYY224 pKa = 11.61ASTSKK229 pKa = 9.13TRR231 pKa = 11.84QQEE234 pKa = 4.11SPARR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84LQKK243 pKa = 10.53SGEE246 pKa = 4.22SPKK249 pKa = 10.38PSSTTSPEE257 pKa = 3.79TRR259 pKa = 11.84SIPLRR264 pKa = 11.84RR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84GEE269 pKa = 4.06GEE271 pKa = 3.99SAEE274 pKa = 4.41GPGGEE279 pKa = 4.41TPTKK283 pKa = 9.59RR284 pKa = 11.84RR285 pKa = 11.84RR286 pKa = 11.84GGRR289 pKa = 11.84EE290 pKa = 3.53LGFVPSPEE298 pKa = 4.3EE299 pKa = 3.45VGSRR303 pKa = 11.84HH304 pKa = 5.23RR305 pKa = 11.84TPEE308 pKa = 3.83TTGLSRR314 pKa = 11.84LGQLQADD321 pKa = 3.63AWDD324 pKa = 3.92PPIILVRR331 pKa = 11.84GLPNTLKK338 pKa = 9.98CWRR341 pKa = 11.84YY342 pKa = 10.14RR343 pKa = 11.84KK344 pKa = 9.08FQNANKK350 pKa = 9.14CFWKK354 pKa = 10.41ISTVFNWVGDD364 pKa = 3.89CDD366 pKa = 3.83KK367 pKa = 11.1SSRR370 pKa = 11.84GRR372 pKa = 11.84LIIAFKK378 pKa = 10.52SDD380 pKa = 2.95KK381 pKa = 10.32QRR383 pKa = 11.84EE384 pKa = 4.5CFVKK388 pKa = 10.15QHH390 pKa = 6.63LFPKK394 pKa = 10.0GSSYY398 pKa = 11.01TYY400 pKa = 10.71GHH402 pKa = 7.22LNGLL406 pKa = 4.02
MM1 pKa = 8.34AMEE4 pKa = 4.65SLTDD8 pKa = 3.98RR9 pKa = 11.84FDD11 pKa = 3.95ALQEE15 pKa = 4.61LILKK19 pKa = 9.93HH20 pKa = 6.32IEE22 pKa = 3.92SDD24 pKa = 3.74STSLQSHH31 pKa = 5.16ITYY34 pKa = 8.85WGYY37 pKa = 10.2VRR39 pKa = 11.84KK40 pKa = 9.81EE41 pKa = 3.7NALMHH46 pKa = 6.2VARR49 pKa = 11.84QQGLTRR55 pKa = 11.84LGLQPLPALPVTEE68 pKa = 4.7YY69 pKa = 10.67NAKK72 pKa = 9.54QAITIQLTLQSLLRR86 pKa = 11.84SQYY89 pKa = 11.21GNEE92 pKa = 3.77PWTLTEE98 pKa = 4.67VSAEE102 pKa = 4.48LINTPPQNLLKK113 pKa = 10.34KK114 pKa = 10.28DD115 pKa = 3.72GFSVEE120 pKa = 3.77VWFDD124 pKa = 3.25NDD126 pKa = 3.08KK127 pKa = 11.13NNAMVYY133 pKa = 8.43TNWNDD138 pKa = 2.96MYY140 pKa = 11.26YY141 pKa = 10.69QDD143 pKa = 5.61AQEE146 pKa = 4.25TWHH149 pKa = 6.63KK150 pKa = 10.43VHH152 pKa = 7.14GSVDD156 pKa = 3.58YY157 pKa = 11.27NGLYY161 pKa = 8.8FTDD164 pKa = 3.91HH165 pKa = 6.19NGEE168 pKa = 3.88RR169 pKa = 11.84AYY171 pKa = 9.7FTLFDD176 pKa = 3.96TDD178 pKa = 4.61AIRR181 pKa = 11.84YY182 pKa = 6.27STTGLWTVHH191 pKa = 5.74YY192 pKa = 8.81KK193 pKa = 10.1SHH195 pKa = 6.57VISSSVVSSTTRR207 pKa = 11.84DD208 pKa = 3.15SSTTPEE214 pKa = 4.3DD215 pKa = 3.7SVDD218 pKa = 3.83RR219 pKa = 11.84PSTSYY224 pKa = 11.61ASTSKK229 pKa = 9.13TRR231 pKa = 11.84QQEE234 pKa = 4.11SPARR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84LQKK243 pKa = 10.53SGEE246 pKa = 4.22SPKK249 pKa = 10.38PSSTTSPEE257 pKa = 3.79TRR259 pKa = 11.84SIPLRR264 pKa = 11.84RR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84GEE269 pKa = 4.06GEE271 pKa = 3.99SAEE274 pKa = 4.41GPGGEE279 pKa = 4.41TPTKK283 pKa = 9.59RR284 pKa = 11.84RR285 pKa = 11.84RR286 pKa = 11.84GGRR289 pKa = 11.84EE290 pKa = 3.53LGFVPSPEE298 pKa = 4.3EE299 pKa = 3.45VGSRR303 pKa = 11.84HH304 pKa = 5.23RR305 pKa = 11.84TPEE308 pKa = 3.83TTGLSRR314 pKa = 11.84LGQLQADD321 pKa = 3.63AWDD324 pKa = 3.92PPIILVRR331 pKa = 11.84GLPNTLKK338 pKa = 9.98CWRR341 pKa = 11.84YY342 pKa = 10.14RR343 pKa = 11.84KK344 pKa = 9.08FQNANKK350 pKa = 9.14CFWKK354 pKa = 10.41ISTVFNWVGDD364 pKa = 3.89CDD366 pKa = 3.83KK367 pKa = 11.1SSRR370 pKa = 11.84GRR372 pKa = 11.84LIIAFKK378 pKa = 10.52SDD380 pKa = 2.95KK381 pKa = 10.32QRR383 pKa = 11.84EE384 pKa = 4.5CFVKK388 pKa = 10.15QHH390 pKa = 6.63LFPKK394 pKa = 10.0GSSYY398 pKa = 11.01TYY400 pKa = 10.71GHH402 pKa = 7.22LNGLL406 pKa = 4.02
Molecular weight: 46.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2277 |
99 |
607 |
379.5 |
42.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.621 ± 0.271 | 2.459 ± 1.039 |
6.28 ± 0.416 | 6.017 ± 0.597 |
4.787 ± 0.437 | 6.061 ± 0.764 |
2.108 ± 0.189 | 5.402 ± 0.572 |
5.094 ± 1.057 | 8.608 ± 0.624 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.581 ± 0.218 | 4.348 ± 0.327 |
5.621 ± 0.943 | 4.48 ± 0.351 |
5.621 ± 0.993 | 7.422 ± 0.795 |
6.807 ± 0.771 | 6.895 ± 0.627 |
1.361 ± 0.359 | 3.426 ± 0.383 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
