
Tritonibacter horizontis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Tritonibacter
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
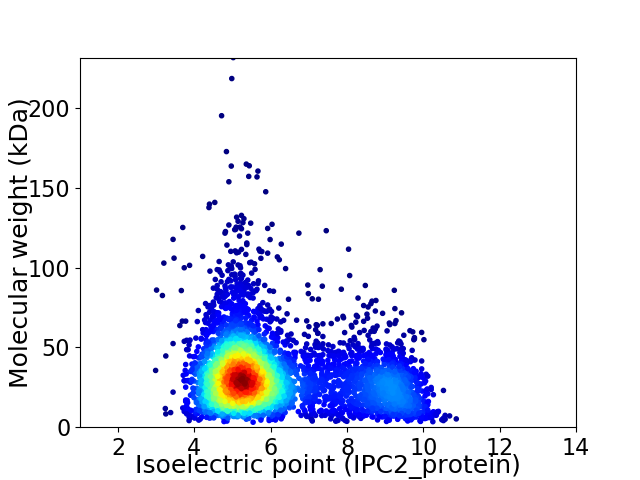
Virtual 2D-PAGE plot for 4579 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
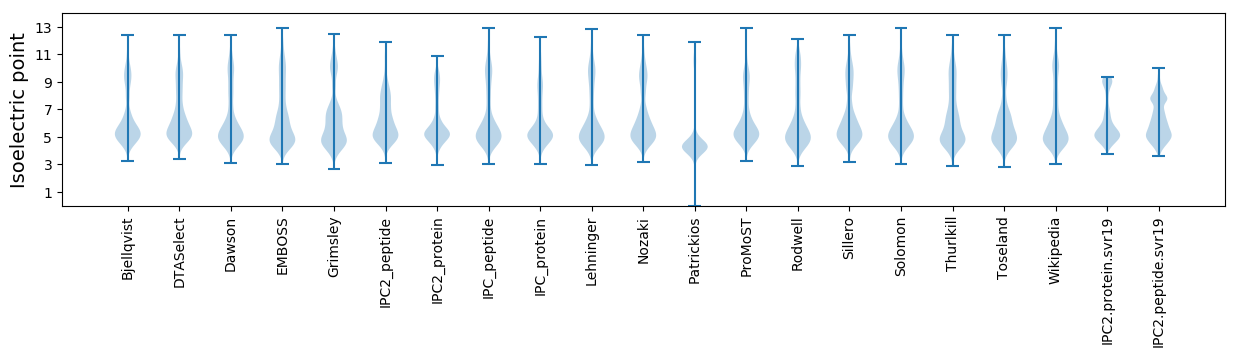
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A132BYW2|A0A132BYW2_9RHOB Galactofuranosyl transferase GlfT2 OS=Tritonibacter horizontis OX=1768241 GN=glfT2_1 PE=4 SV=1
MM1 pKa = 7.84CEE3 pKa = 3.69FCAATTVFDD12 pKa = 4.31PARR15 pKa = 11.84HH16 pKa = 5.9NDD18 pKa = 3.6SEE20 pKa = 4.2GLRR23 pKa = 11.84INGANFFEE31 pKa = 5.69SGDD34 pKa = 3.6ASDD37 pKa = 4.8NLNTGYY43 pKa = 10.54SLPVGGSFYY52 pKa = 11.17GALDD56 pKa = 3.71SVGDD60 pKa = 4.04RR61 pKa = 11.84DD62 pKa = 3.79WVAVTLEE69 pKa = 4.32AGVTYY74 pKa = 10.37QISQSGYY81 pKa = 7.78WGGGGTLYY89 pKa = 10.97DD90 pKa = 3.62GFLRR94 pKa = 11.84VYY96 pKa = 10.36NSNGQQVAGNDD107 pKa = 4.19DD108 pKa = 3.47GRR110 pKa = 11.84HH111 pKa = 5.08TYY113 pKa = 10.17DD114 pKa = 3.84AEE116 pKa = 4.36VNMTVQSGGTFYY128 pKa = 10.71ISVGSYY134 pKa = 10.1RR135 pKa = 11.84DD136 pKa = 3.72SLTGTYY142 pKa = 10.52RR143 pKa = 11.84LDD145 pKa = 3.51IDD147 pKa = 4.55SNAPTVSATEE157 pKa = 3.87EE158 pKa = 3.97AGVQVLADD166 pKa = 4.03YY167 pKa = 10.99LVDD170 pKa = 4.86GYY172 pKa = 10.21WEE174 pKa = 3.82EE175 pKa = 3.97SGRR178 pKa = 11.84DD179 pKa = 3.42EE180 pKa = 5.44RR181 pKa = 11.84SFDD184 pKa = 3.41VSEE187 pKa = 4.5SNEE190 pKa = 3.58ITVNLTGLTAAGQQLARR207 pKa = 11.84WAFEE211 pKa = 3.59AWEE214 pKa = 4.29MVADD218 pKa = 4.22LQFVEE223 pKa = 4.6VSGAAQITYY232 pKa = 10.72SDD234 pKa = 4.47DD235 pKa = 3.43SSGAYY240 pKa = 7.73ATTRR244 pKa = 11.84TWGTEE249 pKa = 3.34ITGVDD254 pKa = 3.39INISQDD260 pKa = 2.19WLTRR264 pKa = 11.84YY265 pKa = 8.02GTEE268 pKa = 3.94LNSYY272 pKa = 10.89SMVAHH277 pKa = 5.05VHH279 pKa = 5.4EE280 pKa = 5.54LGHH283 pKa = 6.66ALGLGHH289 pKa = 6.44QGYY292 pKa = 9.61YY293 pKa = 10.47NGGAVYY299 pKa = 10.38GSNQNFVNDD308 pKa = 3.51SWQMSVMSYY317 pKa = 10.04FSQQEE322 pKa = 4.1NTSVDD327 pKa = 3.07ASYY330 pKa = 11.38GLPITAMMADD340 pKa = 3.41IVAIQSMYY348 pKa = 10.31GAPGADD354 pKa = 3.28AATAGDD360 pKa = 4.37SLWGHH365 pKa = 6.39GSDD368 pKa = 4.47FGNYY372 pKa = 9.61LDD374 pKa = 5.84DD375 pKa = 4.22VFDD378 pKa = 4.27SFLLGGDD385 pKa = 3.37AALGSNRR392 pKa = 11.84NFVFTIYY399 pKa = 10.85DD400 pKa = 3.32SDD402 pKa = 4.76GDD404 pKa = 3.98DD405 pKa = 3.84TLDD408 pKa = 4.02LSSLSTRR415 pKa = 11.84NTVDD419 pKa = 3.36LNGGSFSDD427 pKa = 3.3IQGRR431 pKa = 11.84TGSLAIAEE439 pKa = 4.3DD440 pKa = 4.19TVIEE444 pKa = 3.97NLIMGSGNDD453 pKa = 3.29TVTGNEE459 pKa = 3.69VDD461 pKa = 3.35NHH463 pKa = 6.25IEE465 pKa = 3.86LGGGHH470 pKa = 6.96DD471 pKa = 3.91VAHH474 pKa = 6.92GGAGTDD480 pKa = 4.13RR481 pKa = 11.84IWGGTGTDD489 pKa = 3.74TIYY492 pKa = 11.13GGDD495 pKa = 4.42DD496 pKa = 3.27RR497 pKa = 11.84DD498 pKa = 3.44ILYY501 pKa = 10.84GNSSNDD507 pKa = 3.17VLFGEE512 pKa = 5.14GGNDD516 pKa = 3.52LLVGQHH522 pKa = 6.89HH523 pKa = 7.39NDD525 pKa = 3.48TLNGGLGNDD534 pKa = 3.88VLSGGHH540 pKa = 6.15GADD543 pKa = 3.48VFCFDD548 pKa = 4.94AGSGSDD554 pKa = 3.48RR555 pKa = 11.84IKK557 pKa = 10.92DD558 pKa = 3.67FKK560 pKa = 10.84LWQGDD565 pKa = 3.95SLQLDD570 pKa = 3.54RR571 pKa = 11.84DD572 pKa = 3.83LVEE575 pKa = 6.2DD576 pKa = 4.56EE577 pKa = 5.04LALDD581 pKa = 4.02QLISTYY587 pKa = 11.03GQVTDD592 pKa = 4.07GGLVLTFDD600 pKa = 3.89TGDD603 pKa = 3.31TLEE606 pKa = 4.24FMGIFDD612 pKa = 6.22LDD614 pKa = 3.81SADD617 pKa = 3.87NDD619 pKa = 3.21ILAWVV624 pKa = 3.82
MM1 pKa = 7.84CEE3 pKa = 3.69FCAATTVFDD12 pKa = 4.31PARR15 pKa = 11.84HH16 pKa = 5.9NDD18 pKa = 3.6SEE20 pKa = 4.2GLRR23 pKa = 11.84INGANFFEE31 pKa = 5.69SGDD34 pKa = 3.6ASDD37 pKa = 4.8NLNTGYY43 pKa = 10.54SLPVGGSFYY52 pKa = 11.17GALDD56 pKa = 3.71SVGDD60 pKa = 4.04RR61 pKa = 11.84DD62 pKa = 3.79WVAVTLEE69 pKa = 4.32AGVTYY74 pKa = 10.37QISQSGYY81 pKa = 7.78WGGGGTLYY89 pKa = 10.97DD90 pKa = 3.62GFLRR94 pKa = 11.84VYY96 pKa = 10.36NSNGQQVAGNDD107 pKa = 4.19DD108 pKa = 3.47GRR110 pKa = 11.84HH111 pKa = 5.08TYY113 pKa = 10.17DD114 pKa = 3.84AEE116 pKa = 4.36VNMTVQSGGTFYY128 pKa = 10.71ISVGSYY134 pKa = 10.1RR135 pKa = 11.84DD136 pKa = 3.72SLTGTYY142 pKa = 10.52RR143 pKa = 11.84LDD145 pKa = 3.51IDD147 pKa = 4.55SNAPTVSATEE157 pKa = 3.87EE158 pKa = 3.97AGVQVLADD166 pKa = 4.03YY167 pKa = 10.99LVDD170 pKa = 4.86GYY172 pKa = 10.21WEE174 pKa = 3.82EE175 pKa = 3.97SGRR178 pKa = 11.84DD179 pKa = 3.42EE180 pKa = 5.44RR181 pKa = 11.84SFDD184 pKa = 3.41VSEE187 pKa = 4.5SNEE190 pKa = 3.58ITVNLTGLTAAGQQLARR207 pKa = 11.84WAFEE211 pKa = 3.59AWEE214 pKa = 4.29MVADD218 pKa = 4.22LQFVEE223 pKa = 4.6VSGAAQITYY232 pKa = 10.72SDD234 pKa = 4.47DD235 pKa = 3.43SSGAYY240 pKa = 7.73ATTRR244 pKa = 11.84TWGTEE249 pKa = 3.34ITGVDD254 pKa = 3.39INISQDD260 pKa = 2.19WLTRR264 pKa = 11.84YY265 pKa = 8.02GTEE268 pKa = 3.94LNSYY272 pKa = 10.89SMVAHH277 pKa = 5.05VHH279 pKa = 5.4EE280 pKa = 5.54LGHH283 pKa = 6.66ALGLGHH289 pKa = 6.44QGYY292 pKa = 9.61YY293 pKa = 10.47NGGAVYY299 pKa = 10.38GSNQNFVNDD308 pKa = 3.51SWQMSVMSYY317 pKa = 10.04FSQQEE322 pKa = 4.1NTSVDD327 pKa = 3.07ASYY330 pKa = 11.38GLPITAMMADD340 pKa = 3.41IVAIQSMYY348 pKa = 10.31GAPGADD354 pKa = 3.28AATAGDD360 pKa = 4.37SLWGHH365 pKa = 6.39GSDD368 pKa = 4.47FGNYY372 pKa = 9.61LDD374 pKa = 5.84DD375 pKa = 4.22VFDD378 pKa = 4.27SFLLGGDD385 pKa = 3.37AALGSNRR392 pKa = 11.84NFVFTIYY399 pKa = 10.85DD400 pKa = 3.32SDD402 pKa = 4.76GDD404 pKa = 3.98DD405 pKa = 3.84TLDD408 pKa = 4.02LSSLSTRR415 pKa = 11.84NTVDD419 pKa = 3.36LNGGSFSDD427 pKa = 3.3IQGRR431 pKa = 11.84TGSLAIAEE439 pKa = 4.3DD440 pKa = 4.19TVIEE444 pKa = 3.97NLIMGSGNDD453 pKa = 3.29TVTGNEE459 pKa = 3.69VDD461 pKa = 3.35NHH463 pKa = 6.25IEE465 pKa = 3.86LGGGHH470 pKa = 6.96DD471 pKa = 3.91VAHH474 pKa = 6.92GGAGTDD480 pKa = 4.13RR481 pKa = 11.84IWGGTGTDD489 pKa = 3.74TIYY492 pKa = 11.13GGDD495 pKa = 4.42DD496 pKa = 3.27RR497 pKa = 11.84DD498 pKa = 3.44ILYY501 pKa = 10.84GNSSNDD507 pKa = 3.17VLFGEE512 pKa = 5.14GGNDD516 pKa = 3.52LLVGQHH522 pKa = 6.89HH523 pKa = 7.39NDD525 pKa = 3.48TLNGGLGNDD534 pKa = 3.88VLSGGHH540 pKa = 6.15GADD543 pKa = 3.48VFCFDD548 pKa = 4.94AGSGSDD554 pKa = 3.48RR555 pKa = 11.84IKK557 pKa = 10.92DD558 pKa = 3.67FKK560 pKa = 10.84LWQGDD565 pKa = 3.95SLQLDD570 pKa = 3.54RR571 pKa = 11.84DD572 pKa = 3.83LVEE575 pKa = 6.2DD576 pKa = 4.56EE577 pKa = 5.04LALDD581 pKa = 4.02QLISTYY587 pKa = 11.03GQVTDD592 pKa = 4.07GGLVLTFDD600 pKa = 3.89TGDD603 pKa = 3.31TLEE606 pKa = 4.24FMGIFDD612 pKa = 6.22LDD614 pKa = 3.81SADD617 pKa = 3.87NDD619 pKa = 3.21ILAWVV624 pKa = 3.82
Molecular weight: 66.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A132BUE0|A0A132BUE0_9RHOB Aerobic cobaltochelatase subunit CobN OS=Tritonibacter horizontis OX=1768241 GN=cobN PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 10.3LLDD5 pKa = 3.66IAEE8 pKa = 4.38VAQAAGVTPSTLRR21 pKa = 11.84YY22 pKa = 9.21YY23 pKa = 9.83EE24 pKa = 4.28EE25 pKa = 4.97KK26 pKa = 11.17GLIQPVARR34 pKa = 11.84HH35 pKa = 5.12GLRR38 pKa = 11.84RR39 pKa = 11.84QYY41 pKa = 11.0GPQVLQQLSLVAMGKK56 pKa = 8.33MAGFSLDD63 pKa = 2.9EE64 pKa = 4.12VAAMFGPTGRR74 pKa = 11.84PDD76 pKa = 3.21MPRR79 pKa = 11.84NDD81 pKa = 3.88LLRR84 pKa = 11.84RR85 pKa = 11.84AEE87 pKa = 4.03RR88 pKa = 11.84MQAQARR94 pKa = 11.84QLDD97 pKa = 3.9ALAQMLRR104 pKa = 11.84HH105 pKa = 5.43VAEE108 pKa = 5.13CPAPTHH114 pKa = 6.62LDD116 pKa = 3.22CDD118 pKa = 3.81KK119 pKa = 10.82FQQLLRR125 pKa = 11.84LATATQKK132 pKa = 10.16RR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84SGQSPSRR143 pKa = 3.69
MM1 pKa = 7.6KK2 pKa = 10.3LLDD5 pKa = 3.66IAEE8 pKa = 4.38VAQAAGVTPSTLRR21 pKa = 11.84YY22 pKa = 9.21YY23 pKa = 9.83EE24 pKa = 4.28EE25 pKa = 4.97KK26 pKa = 11.17GLIQPVARR34 pKa = 11.84HH35 pKa = 5.12GLRR38 pKa = 11.84RR39 pKa = 11.84QYY41 pKa = 11.0GPQVLQQLSLVAMGKK56 pKa = 8.33MAGFSLDD63 pKa = 2.9EE64 pKa = 4.12VAAMFGPTGRR74 pKa = 11.84PDD76 pKa = 3.21MPRR79 pKa = 11.84NDD81 pKa = 3.88LLRR84 pKa = 11.84RR85 pKa = 11.84AEE87 pKa = 4.03RR88 pKa = 11.84MQAQARR94 pKa = 11.84QLDD97 pKa = 3.9ALAQMLRR104 pKa = 11.84HH105 pKa = 5.43VAEE108 pKa = 5.13CPAPTHH114 pKa = 6.62LDD116 pKa = 3.22CDD118 pKa = 3.81KK119 pKa = 10.82FQQLLRR125 pKa = 11.84LATATQKK132 pKa = 10.16RR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84SGQSPSRR143 pKa = 3.69
Molecular weight: 16.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1430001 |
29 |
2153 |
312.3 |
33.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.25 ± 0.045 | 0.927 ± 0.01 |
6.088 ± 0.032 | 5.805 ± 0.034 |
3.736 ± 0.022 | 8.436 ± 0.035 |
2.067 ± 0.018 | 5.079 ± 0.027 |
3.126 ± 0.03 | 10.306 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.73 ± 0.016 | 2.602 ± 0.021 |
4.992 ± 0.027 | 3.508 ± 0.021 |
6.677 ± 0.036 | 5.358 ± 0.022 |
5.546 ± 0.022 | 7.213 ± 0.029 |
1.344 ± 0.015 | 2.211 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
