
Klebsiella pneumoniae subsp. ozaenae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Klebsiella; Klebsiella pneumoniae
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
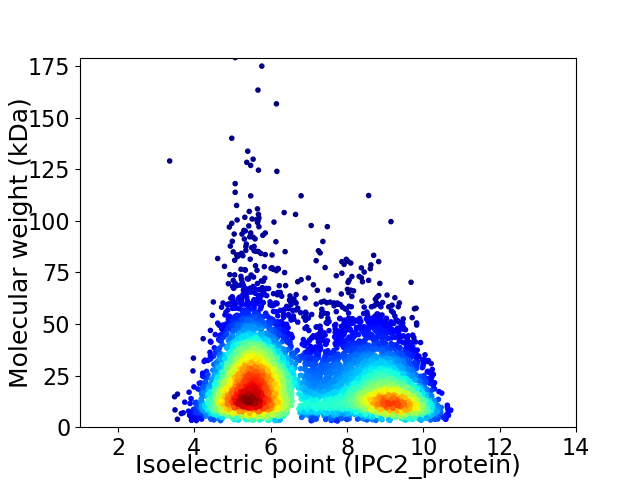
Virtual 2D-PAGE plot for 6726 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A377Z5S6|A0A377Z5S6_KLEPO Periplasmic hemin-binding protein OS=Klebsiella pneumoniae subsp. ozaenae OX=574 GN=hmuT_1 PE=4 SV=1
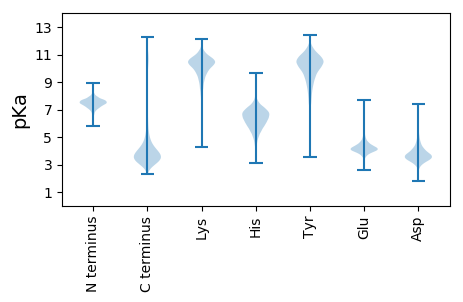
MM1 pKa = 7.67NIGLFYY7 pKa = 10.91GSSTCYY13 pKa = 10.49TEE15 pKa = 3.84MAAEE19 pKa = 5.12KK20 pKa = 10.0IRR22 pKa = 11.84DD23 pKa = 3.65IIGPEE28 pKa = 4.02LVTLHH33 pKa = 6.12NLKK36 pKa = 10.49DD37 pKa = 3.83DD38 pKa = 4.23SPALMSQYY46 pKa = 11.27DD47 pKa = 3.97VLILGIPTWDD57 pKa = 3.26FGEE60 pKa = 4.22IQEE63 pKa = 4.44DD64 pKa = 4.06WEE66 pKa = 4.5AVWDD70 pKa = 4.34QLDD73 pKa = 3.73TLNLEE78 pKa = 4.43GKK80 pKa = 9.65IVALYY85 pKa = 11.09GMGDD89 pKa = 3.29QLGYY93 pKa = 11.02GEE95 pKa = 4.87WFLDD99 pKa = 3.74ALGMLHH105 pKa = 7.54DD106 pKa = 4.49KK107 pKa = 10.83LATKK111 pKa = 10.04GVKK114 pKa = 9.5FVGYY118 pKa = 9.0WPTEE122 pKa = 4.03GYY124 pKa = 10.61EE125 pKa = 4.19FTSPKK130 pKa = 10.02PVIADD135 pKa = 3.41GQLFVGLALDD145 pKa = 3.64EE146 pKa = 4.38TNQYY150 pKa = 10.89DD151 pKa = 4.3LSDD154 pKa = 3.61EE155 pKa = 5.41RR156 pKa = 11.84IQSWCEE162 pKa = 3.63QILGEE167 pKa = 4.17MAEE170 pKa = 4.4HH171 pKa = 7.09FSS173 pKa = 3.73
MM1 pKa = 7.67NIGLFYY7 pKa = 10.91GSSTCYY13 pKa = 10.49TEE15 pKa = 3.84MAAEE19 pKa = 5.12KK20 pKa = 10.0IRR22 pKa = 11.84DD23 pKa = 3.65IIGPEE28 pKa = 4.02LVTLHH33 pKa = 6.12NLKK36 pKa = 10.49DD37 pKa = 3.83DD38 pKa = 4.23SPALMSQYY46 pKa = 11.27DD47 pKa = 3.97VLILGIPTWDD57 pKa = 3.26FGEE60 pKa = 4.22IQEE63 pKa = 4.44DD64 pKa = 4.06WEE66 pKa = 4.5AVWDD70 pKa = 4.34QLDD73 pKa = 3.73TLNLEE78 pKa = 4.43GKK80 pKa = 9.65IVALYY85 pKa = 11.09GMGDD89 pKa = 3.29QLGYY93 pKa = 11.02GEE95 pKa = 4.87WFLDD99 pKa = 3.74ALGMLHH105 pKa = 7.54DD106 pKa = 4.49KK107 pKa = 10.83LATKK111 pKa = 10.04GVKK114 pKa = 9.5FVGYY118 pKa = 9.0WPTEE122 pKa = 4.03GYY124 pKa = 10.61EE125 pKa = 4.19FTSPKK130 pKa = 10.02PVIADD135 pKa = 3.41GQLFVGLALDD145 pKa = 3.64EE146 pKa = 4.38TNQYY150 pKa = 10.89DD151 pKa = 4.3LSDD154 pKa = 3.61EE155 pKa = 5.41RR156 pKa = 11.84IQSWCEE162 pKa = 3.63QILGEE167 pKa = 4.17MAEE170 pKa = 4.4HH171 pKa = 7.09FSS173 pKa = 3.73
Molecular weight: 19.51 kDa
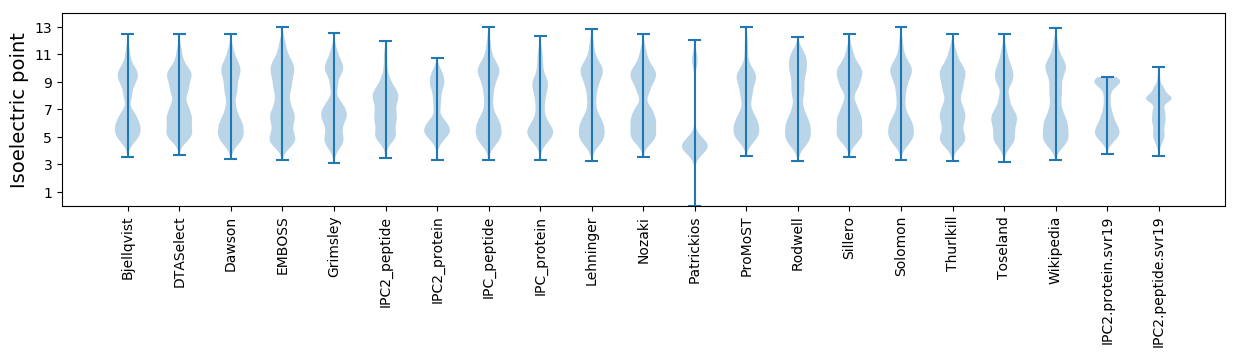
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A378BX62|A0A378BX62_KLEPO Branched-chain amino acid transport system carrier protein OS=Klebsiella pneumoniae subsp. ozaenae OX=574 GN=brnQ PE=3 SV=1
MM1 pKa = 7.42FGGKK5 pKa = 10.0GGLGNLMKK13 pKa = 10.29QAQQMQEE20 pKa = 3.99KK21 pKa = 7.38MQKK24 pKa = 7.25MQEE27 pKa = 4.08EE28 pKa = 4.47IAQLEE33 pKa = 4.52VTGEE37 pKa = 4.05SGAGLVKK44 pKa = 9.63VTINGAHH51 pKa = 6.01NCRR54 pKa = 11.84RR55 pKa = 11.84VEE57 pKa = 4.1SRR59 pKa = 11.84PKK61 pKa = 9.42PAGRR65 pKa = 11.84RR66 pKa = 11.84QRR68 pKa = 11.84DD69 pKa = 3.15ARR71 pKa = 11.84RR72 pKa = 11.84SGGCRR77 pKa = 11.84VQRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84APYY85 pKa = 9.25RR86 pKa = 11.84RR87 pKa = 11.84DD88 pKa = 3.38PEE90 pKa = 4.71RR91 pKa = 11.84EE92 pKa = 3.71DD93 pKa = 3.28GFRR96 pKa = 11.84FCRR99 pKa = 11.84YY100 pKa = 7.45ATAARR105 pKa = 11.84LQDD108 pKa = 3.66AVLMQTSPLLTQLMEE123 pKa = 4.94ALRR126 pKa = 11.84CLPGVGPKK134 pKa = 9.64SAQRR138 pKa = 11.84MAFTLLQRR146 pKa = 11.84DD147 pKa = 3.41RR148 pKa = 11.84SGGMRR153 pKa = 11.84LAQALTRR160 pKa = 11.84AMSKK164 pKa = 9.73IGHH167 pKa = 6.56CADD170 pKa = 3.21CRR172 pKa = 11.84TFTEE176 pKa = 4.26QEE178 pKa = 3.91VCNICSNPRR187 pKa = 11.84RR188 pKa = 11.84QEE190 pKa = 3.59NGQICVVEE198 pKa = 4.71SPADD202 pKa = 3.14IYY204 pKa = 11.28AIEE207 pKa = 4.11QTGQYY212 pKa = 8.99SGRR215 pKa = 11.84YY216 pKa = 7.12FVLMGTCRR224 pKa = 11.84RR225 pKa = 11.84STASGLMTSGSIAWSNGWRR244 pKa = 11.84RR245 pKa = 11.84SRR247 pKa = 11.84SPRR250 pKa = 3.28
MM1 pKa = 7.42FGGKK5 pKa = 10.0GGLGNLMKK13 pKa = 10.29QAQQMQEE20 pKa = 3.99KK21 pKa = 7.38MQKK24 pKa = 7.25MQEE27 pKa = 4.08EE28 pKa = 4.47IAQLEE33 pKa = 4.52VTGEE37 pKa = 4.05SGAGLVKK44 pKa = 9.63VTINGAHH51 pKa = 6.01NCRR54 pKa = 11.84RR55 pKa = 11.84VEE57 pKa = 4.1SRR59 pKa = 11.84PKK61 pKa = 9.42PAGRR65 pKa = 11.84RR66 pKa = 11.84QRR68 pKa = 11.84DD69 pKa = 3.15ARR71 pKa = 11.84RR72 pKa = 11.84SGGCRR77 pKa = 11.84VQRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84APYY85 pKa = 9.25RR86 pKa = 11.84RR87 pKa = 11.84DD88 pKa = 3.38PEE90 pKa = 4.71RR91 pKa = 11.84EE92 pKa = 3.71DD93 pKa = 3.28GFRR96 pKa = 11.84FCRR99 pKa = 11.84YY100 pKa = 7.45ATAARR105 pKa = 11.84LQDD108 pKa = 3.66AVLMQTSPLLTQLMEE123 pKa = 4.94ALRR126 pKa = 11.84CLPGVGPKK134 pKa = 9.64SAQRR138 pKa = 11.84MAFTLLQRR146 pKa = 11.84DD147 pKa = 3.41RR148 pKa = 11.84SGGMRR153 pKa = 11.84LAQALTRR160 pKa = 11.84AMSKK164 pKa = 9.73IGHH167 pKa = 6.56CADD170 pKa = 3.21CRR172 pKa = 11.84TFTEE176 pKa = 4.26QEE178 pKa = 3.91VCNICSNPRR187 pKa = 11.84RR188 pKa = 11.84QEE190 pKa = 3.59NGQICVVEE198 pKa = 4.71SPADD202 pKa = 3.14IYY204 pKa = 11.28AIEE207 pKa = 4.11QTGQYY212 pKa = 8.99SGRR215 pKa = 11.84YY216 pKa = 7.12FVLMGTCRR224 pKa = 11.84RR225 pKa = 11.84STASGLMTSGSIAWSNGWRR244 pKa = 11.84RR245 pKa = 11.84SRR247 pKa = 11.84SPRR250 pKa = 3.28
Molecular weight: 27.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1452714 |
29 |
1649 |
216.0 |
23.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.423 ± 0.044 | 1.157 ± 0.012 |
5.061 ± 0.025 | 5.364 ± 0.034 |
3.746 ± 0.023 | 7.646 ± 0.036 |
2.316 ± 0.017 | 5.461 ± 0.027 |
3.847 ± 0.028 | 10.908 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.81 ± 0.016 | 3.356 ± 0.021 |
4.763 ± 0.024 | 4.512 ± 0.032 |
6.385 ± 0.035 | 5.835 ± 0.029 |
5.144 ± 0.024 | 6.96 ± 0.027 |
1.605 ± 0.013 | 2.702 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
