
Saprochaete ingens
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Dipodascaceae; Saprochaete
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
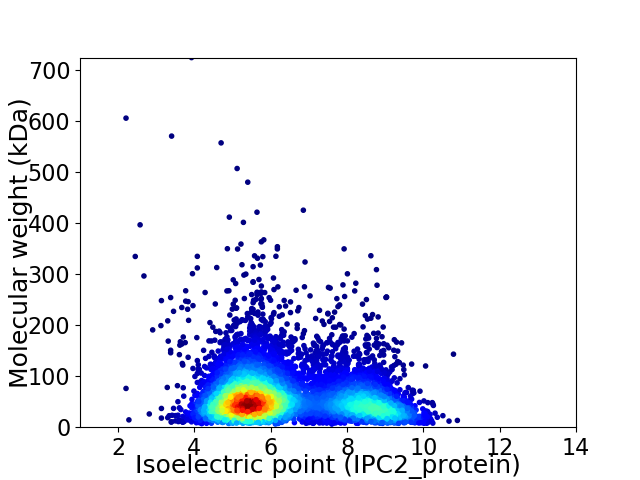
Virtual 2D-PAGE plot for 6465 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5E8BHE6|A0A5E8BHE6_9ASCO Uncharacterized protein OS=Saprochaete ingens OX=2606893 GN=SAPINGB_P002975 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.38FSDD5 pKa = 3.56ILFSTSAVMMVNALPSNILFPKK27 pKa = 9.97RR28 pKa = 11.84QVLEE32 pKa = 4.42DD33 pKa = 3.6YY34 pKa = 11.04NIISVQGSSSTSTLSAVPTSSLNSTSQYY62 pKa = 11.21NLNNSTATLNSTLLQNSTTTTLAPYY87 pKa = 9.37TNATVVVTEE96 pKa = 4.84FGTPVTKK103 pKa = 9.92TVLSIVTQVVIVDD116 pKa = 3.43SDD118 pKa = 4.16GNVITGEE125 pKa = 4.1YY126 pKa = 9.82NDD128 pKa = 4.65AGANEE133 pKa = 4.11NSVAASVSSGAGSQVVTVSVDD154 pKa = 3.22AASAAATSSSDD165 pKa = 3.35SSSSSSTSVSAGSDD179 pKa = 3.17TSSSNGSSDD188 pKa = 3.23SSGSNSSSGSSSTSNSSGSSDD209 pKa = 3.23SSSNSLGSSDD219 pKa = 4.23SLSSSSSSSSDD230 pKa = 3.14SSSSSSNTSSGTSSSSSGSGSGSSEE255 pKa = 3.69TSANTSSSSSSASSDD270 pKa = 3.18SSSTNNEE277 pKa = 3.78SSGSSGSSSNTGSSSSPLATLSANLQLVPVSSSTSTSSSDD317 pKa = 3.15EE318 pKa = 4.12STKK321 pKa = 10.93SSVIEE326 pKa = 3.99STISSSSSSSSGSNAIEE343 pKa = 4.31SFSSSGSNTILSSSGSNTIEE363 pKa = 4.13SSSSSSTSTSVSSFSSDD380 pKa = 2.95SPFFVAVTTILPGSATIPTSSPASPSSPSSSSSTSPSLASNADD423 pKa = 3.18SAAEE427 pKa = 3.95ITSKK431 pKa = 10.64ISGLVSSSSSSIITHH446 pKa = 5.48SAYY449 pKa = 11.06LNATVTTAASTASTISSNSTVLAHH473 pKa = 6.85SISSSTATALSTSTFSTLNATMEE496 pKa = 4.16FALKK500 pKa = 8.71HH501 pKa = 4.64THH503 pKa = 5.87SASATSATSAATTTAPASSTAAPSLSKK530 pKa = 10.7LVEE533 pKa = 4.0TSKK536 pKa = 10.13TNVSTVSSSATDD548 pKa = 3.44EE549 pKa = 4.29SSSFSAVTLSLGSIVAANFSTEE571 pKa = 3.66ISGSADD577 pKa = 2.85KK578 pKa = 10.23STASISSVEE587 pKa = 4.03YY588 pKa = 9.59STSLSDD594 pKa = 4.13ASVTEE599 pKa = 4.19SAVASAIEE607 pKa = 4.14SSVEE611 pKa = 3.79STIASSVSSSAEE623 pKa = 3.85VPTEE627 pKa = 3.57SSYY630 pKa = 11.21ISSSEE635 pKa = 4.07SSSAAPIVSSVEE647 pKa = 4.03SSVDD651 pKa = 3.22PTTEE655 pKa = 3.58SSVISEE661 pKa = 4.19STAISSSSTDD671 pKa = 3.12LSSFFSVEE679 pKa = 4.01SSVEE683 pKa = 3.83SDD685 pKa = 4.38PISSTSVSSSSSSSSFSATTIDD707 pKa = 3.58SLEE710 pKa = 4.27SFTDD714 pKa = 3.5AASVNVANKK723 pKa = 9.6QNEE726 pKa = 4.08ITIVATDD733 pKa = 3.57SSSASSSSSSSSSSSEE749 pKa = 3.99WIVSAATLSVNAVEE763 pKa = 4.92PSAAQPSSSTADD775 pKa = 3.6PQSSTGAASTAADD788 pKa = 3.43SAADD792 pKa = 3.54VVVALGAGATDD803 pKa = 3.77AAGQVINTVQQQTSVSVQTVNFDD826 pKa = 3.47PTTTSSSSSSAVVVVVEE843 pKa = 4.62TPVQEE848 pKa = 4.51AQTSTSSTWVPEE860 pKa = 3.99AVPTTSSTTYY870 pKa = 9.66TPEE873 pKa = 4.01AVPTTSSTSTPEE885 pKa = 3.88AVPTTSSTSTPDD897 pKa = 3.3AVPTTSTTSTPDD909 pKa = 3.21AVPTTSSQVIATTSSTAAAASTTTSTSSDD938 pKa = 2.35GWTDD942 pKa = 3.14YY943 pKa = 11.08DD944 pKa = 4.16YY945 pKa = 11.39PGDD948 pKa = 4.56DD949 pKa = 4.16GLFKK953 pKa = 9.87FTPRR957 pKa = 11.84SLSYY961 pKa = 10.91TPFNNDD967 pKa = 2.67TSCRR971 pKa = 11.84STSEE975 pKa = 3.7IAQDD979 pKa = 5.36LADD982 pKa = 5.33LYY984 pKa = 11.49NKK986 pKa = 10.1GIRR989 pKa = 11.84EE990 pKa = 4.02LRR992 pKa = 11.84LYY994 pKa = 10.81GADD997 pKa = 3.39CDD999 pKa = 4.77AINTILSPAKK1009 pKa = 10.27AQGLTVIMGFYY1020 pKa = 8.45VTSAGVDD1027 pKa = 3.62VVDD1030 pKa = 4.83SEE1032 pKa = 4.74VEE1034 pKa = 4.3SFISWAQSSGDD1045 pKa = 3.4IDD1047 pKa = 3.99VVTAVAIGNEE1057 pKa = 3.99AVTNGWVTADD1067 pKa = 3.82DD1068 pKa = 4.55LASKK1072 pKa = 9.85MEE1074 pKa = 4.25SVRR1077 pKa = 11.84SKK1079 pKa = 10.42LQSEE1083 pKa = 4.31IGFTGPVITSEE1094 pKa = 3.86IVGIYY1099 pKa = 9.41QSYY1102 pKa = 8.75PQLCTSSAADD1112 pKa = 3.35YY1113 pKa = 10.8VGVNIHH1119 pKa = 7.1PYY1121 pKa = 9.4FDD1123 pKa = 3.5SGIAADD1129 pKa = 3.71QSGDD1133 pKa = 3.31FMLAQVQLVKK1143 pKa = 10.51DD1144 pKa = 3.28ACGRR1148 pKa = 11.84SDD1150 pKa = 3.73VKK1152 pKa = 11.13VLEE1155 pKa = 4.03TGYY1158 pKa = 9.83PHH1160 pKa = 7.93DD1161 pKa = 4.83GNTNGLQVPSDD1172 pKa = 4.1VNQGIAVTQIYY1183 pKa = 10.01NALQGDD1189 pKa = 4.46VYY1191 pKa = 10.04MFSMWDD1197 pKa = 3.7DD1198 pKa = 3.03HH1199 pKa = 7.02WKK1201 pKa = 10.63DD1202 pKa = 3.1AGNYY1206 pKa = 7.65NVEE1209 pKa = 3.97WYY1211 pKa = 10.38FGLFYY1216 pKa = 10.85RR1217 pKa = 11.84FPP1219 pKa = 5.33
MM1 pKa = 7.48KK2 pKa = 10.38FSDD5 pKa = 3.56ILFSTSAVMMVNALPSNILFPKK27 pKa = 9.97RR28 pKa = 11.84QVLEE32 pKa = 4.42DD33 pKa = 3.6YY34 pKa = 11.04NIISVQGSSSTSTLSAVPTSSLNSTSQYY62 pKa = 11.21NLNNSTATLNSTLLQNSTTTTLAPYY87 pKa = 9.37TNATVVVTEE96 pKa = 4.84FGTPVTKK103 pKa = 9.92TVLSIVTQVVIVDD116 pKa = 3.43SDD118 pKa = 4.16GNVITGEE125 pKa = 4.1YY126 pKa = 9.82NDD128 pKa = 4.65AGANEE133 pKa = 4.11NSVAASVSSGAGSQVVTVSVDD154 pKa = 3.22AASAAATSSSDD165 pKa = 3.35SSSSSSTSVSAGSDD179 pKa = 3.17TSSSNGSSDD188 pKa = 3.23SSGSNSSSGSSSTSNSSGSSDD209 pKa = 3.23SSSNSLGSSDD219 pKa = 4.23SLSSSSSSSSDD230 pKa = 3.14SSSSSSNTSSGTSSSSSGSGSGSSEE255 pKa = 3.69TSANTSSSSSSASSDD270 pKa = 3.18SSSTNNEE277 pKa = 3.78SSGSSGSSSNTGSSSSPLATLSANLQLVPVSSSTSTSSSDD317 pKa = 3.15EE318 pKa = 4.12STKK321 pKa = 10.93SSVIEE326 pKa = 3.99STISSSSSSSSGSNAIEE343 pKa = 4.31SFSSSGSNTILSSSGSNTIEE363 pKa = 4.13SSSSSSTSTSVSSFSSDD380 pKa = 2.95SPFFVAVTTILPGSATIPTSSPASPSSPSSSSSTSPSLASNADD423 pKa = 3.18SAAEE427 pKa = 3.95ITSKK431 pKa = 10.64ISGLVSSSSSSIITHH446 pKa = 5.48SAYY449 pKa = 11.06LNATVTTAASTASTISSNSTVLAHH473 pKa = 6.85SISSSTATALSTSTFSTLNATMEE496 pKa = 4.16FALKK500 pKa = 8.71HH501 pKa = 4.64THH503 pKa = 5.87SASATSATSAATTTAPASSTAAPSLSKK530 pKa = 10.7LVEE533 pKa = 4.0TSKK536 pKa = 10.13TNVSTVSSSATDD548 pKa = 3.44EE549 pKa = 4.29SSSFSAVTLSLGSIVAANFSTEE571 pKa = 3.66ISGSADD577 pKa = 2.85KK578 pKa = 10.23STASISSVEE587 pKa = 4.03YY588 pKa = 9.59STSLSDD594 pKa = 4.13ASVTEE599 pKa = 4.19SAVASAIEE607 pKa = 4.14SSVEE611 pKa = 3.79STIASSVSSSAEE623 pKa = 3.85VPTEE627 pKa = 3.57SSYY630 pKa = 11.21ISSSEE635 pKa = 4.07SSSAAPIVSSVEE647 pKa = 4.03SSVDD651 pKa = 3.22PTTEE655 pKa = 3.58SSVISEE661 pKa = 4.19STAISSSSTDD671 pKa = 3.12LSSFFSVEE679 pKa = 4.01SSVEE683 pKa = 3.83SDD685 pKa = 4.38PISSTSVSSSSSSSSFSATTIDD707 pKa = 3.58SLEE710 pKa = 4.27SFTDD714 pKa = 3.5AASVNVANKK723 pKa = 9.6QNEE726 pKa = 4.08ITIVATDD733 pKa = 3.57SSSASSSSSSSSSSSEE749 pKa = 3.99WIVSAATLSVNAVEE763 pKa = 4.92PSAAQPSSSTADD775 pKa = 3.6PQSSTGAASTAADD788 pKa = 3.43SAADD792 pKa = 3.54VVVALGAGATDD803 pKa = 3.77AAGQVINTVQQQTSVSVQTVNFDD826 pKa = 3.47PTTTSSSSSSAVVVVVEE843 pKa = 4.62TPVQEE848 pKa = 4.51AQTSTSSTWVPEE860 pKa = 3.99AVPTTSSTTYY870 pKa = 9.66TPEE873 pKa = 4.01AVPTTSSTSTPEE885 pKa = 3.88AVPTTSSTSTPDD897 pKa = 3.3AVPTTSTTSTPDD909 pKa = 3.21AVPTTSSQVIATTSSTAAAASTTTSTSSDD938 pKa = 2.35GWTDD942 pKa = 3.14YY943 pKa = 11.08DD944 pKa = 4.16YY945 pKa = 11.39PGDD948 pKa = 4.56DD949 pKa = 4.16GLFKK953 pKa = 9.87FTPRR957 pKa = 11.84SLSYY961 pKa = 10.91TPFNNDD967 pKa = 2.67TSCRR971 pKa = 11.84STSEE975 pKa = 3.7IAQDD979 pKa = 5.36LADD982 pKa = 5.33LYY984 pKa = 11.49NKK986 pKa = 10.1GIRR989 pKa = 11.84EE990 pKa = 4.02LRR992 pKa = 11.84LYY994 pKa = 10.81GADD997 pKa = 3.39CDD999 pKa = 4.77AINTILSPAKK1009 pKa = 10.27AQGLTVIMGFYY1020 pKa = 8.45VTSAGVDD1027 pKa = 3.62VVDD1030 pKa = 4.83SEE1032 pKa = 4.74VEE1034 pKa = 4.3SFISWAQSSGDD1045 pKa = 3.4IDD1047 pKa = 3.99VVTAVAIGNEE1057 pKa = 3.99AVTNGWVTADD1067 pKa = 3.82DD1068 pKa = 4.55LASKK1072 pKa = 9.85MEE1074 pKa = 4.25SVRR1077 pKa = 11.84SKK1079 pKa = 10.42LQSEE1083 pKa = 4.31IGFTGPVITSEE1094 pKa = 3.86IVGIYY1099 pKa = 9.41QSYY1102 pKa = 8.75PQLCTSSAADD1112 pKa = 3.35YY1113 pKa = 10.8VGVNIHH1119 pKa = 7.1PYY1121 pKa = 9.4FDD1123 pKa = 3.5SGIAADD1129 pKa = 3.71QSGDD1133 pKa = 3.31FMLAQVQLVKK1143 pKa = 10.51DD1144 pKa = 3.28ACGRR1148 pKa = 11.84SDD1150 pKa = 3.73VKK1152 pKa = 11.13VLEE1155 pKa = 4.03TGYY1158 pKa = 9.83PHH1160 pKa = 7.93DD1161 pKa = 4.83GNTNGLQVPSDD1172 pKa = 4.1VNQGIAVTQIYY1183 pKa = 10.01NALQGDD1189 pKa = 4.46VYY1191 pKa = 10.04MFSMWDD1197 pKa = 3.7DD1198 pKa = 3.03HH1199 pKa = 7.02WKK1201 pKa = 10.63DD1202 pKa = 3.1AGNYY1206 pKa = 7.65NVEE1209 pKa = 3.97WYY1211 pKa = 10.38FGLFYY1216 pKa = 10.85RR1217 pKa = 11.84FPP1219 pKa = 5.33
Molecular weight: 121.78 kDa
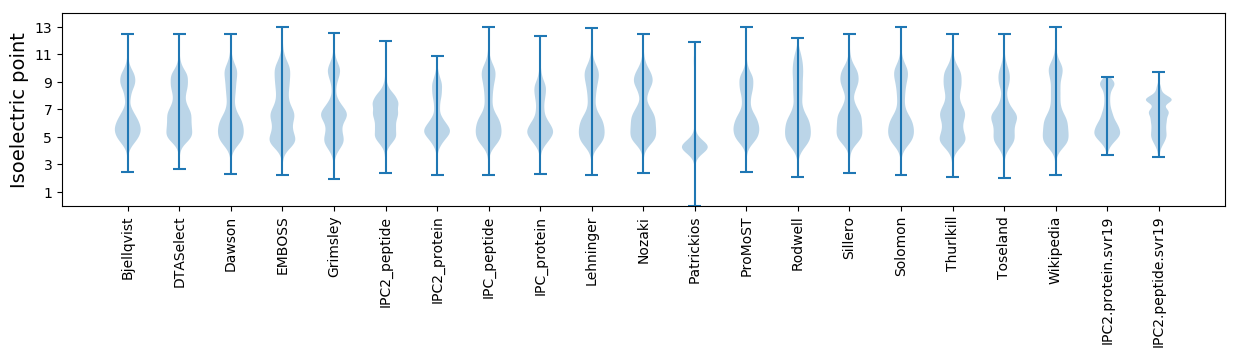
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5E8BV45|A0A5E8BV45_9ASCO Uncharacterized protein OS=Saprochaete ingens OX=2606893 GN=SAPINGB_P004359 PE=4 SV=1
MM1 pKa = 7.82LSRR4 pKa = 11.84FAAIRR9 pKa = 11.84PAMRR13 pKa = 11.84VVASGVASRR22 pKa = 11.84PLTLLAAQSNPLTALRR38 pKa = 11.84ARR40 pKa = 11.84MTPGSPTAALVSPLAQPNPLLLPALPDD67 pKa = 3.81LLDD70 pKa = 3.32QRR72 pKa = 11.84RR73 pKa = 11.84WKK75 pKa = 10.92ARR77 pKa = 11.84GNTYY81 pKa = 10.27QPSTLKK87 pKa = 10.36RR88 pKa = 11.84KK89 pKa = 10.0RR90 pKa = 11.84INGFLARR97 pKa = 11.84LKK99 pKa = 9.8TRR101 pKa = 11.84GGRR104 pKa = 11.84KK105 pKa = 8.7VLARR109 pKa = 11.84RR110 pKa = 11.84KK111 pKa = 8.08TKK113 pKa = 10.28GRR115 pKa = 11.84WFLSHH120 pKa = 7.05
MM1 pKa = 7.82LSRR4 pKa = 11.84FAAIRR9 pKa = 11.84PAMRR13 pKa = 11.84VVASGVASRR22 pKa = 11.84PLTLLAAQSNPLTALRR38 pKa = 11.84ARR40 pKa = 11.84MTPGSPTAALVSPLAQPNPLLLPALPDD67 pKa = 3.81LLDD70 pKa = 3.32QRR72 pKa = 11.84RR73 pKa = 11.84WKK75 pKa = 10.92ARR77 pKa = 11.84GNTYY81 pKa = 10.27QPSTLKK87 pKa = 10.36RR88 pKa = 11.84KK89 pKa = 10.0RR90 pKa = 11.84INGFLARR97 pKa = 11.84LKK99 pKa = 9.8TRR101 pKa = 11.84GGRR104 pKa = 11.84KK105 pKa = 8.7VLARR109 pKa = 11.84RR110 pKa = 11.84KK111 pKa = 8.08TKK113 pKa = 10.28GRR115 pKa = 11.84WFLSHH120 pKa = 7.05
Molecular weight: 13.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3935327 |
66 |
7175 |
608.7 |
67.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.571 ± 0.062 | 0.964 ± 0.011 |
5.465 ± 0.033 | 6.217 ± 0.042 |
3.79 ± 0.023 | 5.688 ± 0.039 |
2.294 ± 0.016 | 5.418 ± 0.025 |
5.533 ± 0.036 | 8.431 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.752 ± 0.013 | 4.94 ± 0.028 |
5.605 ± 0.037 | 4.549 ± 0.046 |
4.665 ± 0.03 | 10.831 ± 0.108 |
6.505 ± 0.036 | 5.875 ± 0.028 |
0.941 ± 0.009 | 2.966 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
