
Collimonas fungivorans (strain Ter331)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Oxalobacteraceae; Collimonas; Collimonas fungivorans
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
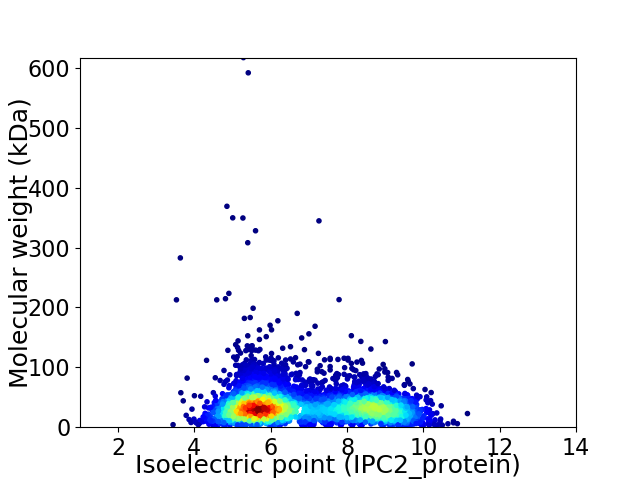
Virtual 2D-PAGE plot for 4429 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G0AGS4|G0AGS4_COLFT Lipoprotein putative OS=Collimonas fungivorans (strain Ter331) OX=1005048 GN=CFU_1855 PE=4 SV=1
MM1 pKa = 7.87TDD3 pKa = 2.78THH5 pKa = 6.7TPSQMPFDD13 pKa = 4.78AGQLQKK19 pKa = 10.45IRR21 pKa = 11.84TLLKK25 pKa = 10.31QQGLLDD31 pKa = 4.22ANGKK35 pKa = 9.36VVDD38 pKa = 4.07TLATFGNSPPKK49 pKa = 10.53GAFAPIYY56 pKa = 10.72AEE58 pKa = 3.5ISKK61 pKa = 10.29IISAVGSTVDD71 pKa = 4.3PKK73 pKa = 9.95IQYY76 pKa = 8.01WFSKK80 pKa = 10.58AADD83 pKa = 3.54INNDD87 pKa = 3.35VTSDD91 pKa = 3.62PADD94 pKa = 3.88DD95 pKa = 4.35YY96 pKa = 11.72IRR98 pKa = 11.84GVTQYY103 pKa = 11.07GLAMRR108 pKa = 11.84HH109 pKa = 5.9KK110 pKa = 10.86LDD112 pKa = 4.24GLSQSAIDD120 pKa = 4.51IGLQQTSNLIGEE132 pKa = 4.3NVINEE137 pKa = 3.91ILGDD141 pKa = 3.68GGIPDD146 pKa = 4.71FGYY149 pKa = 9.81MISGDD154 pKa = 3.03ISAAIGNDD162 pKa = 2.9ALGRR166 pKa = 11.84FQQDD170 pKa = 2.43AGGWGGSFYY179 pKa = 11.21YY180 pKa = 10.15WNAPLNYY187 pKa = 8.05GTTLPVISVGNVIANGGTVLTNGAAVTYY215 pKa = 9.23PSSLKK220 pKa = 10.14EE221 pKa = 3.55FYY223 pKa = 10.09YY224 pKa = 11.22ANSNASADD232 pKa = 3.65FAIPEE237 pKa = 4.15ISTVAASSSVGGILGTLITQGGLPNFPKK265 pKa = 10.45AVNDD269 pKa = 3.04AWVAVLSGSQAQTPIMVKK287 pKa = 10.58AKK289 pKa = 10.47LFADD293 pKa = 4.66TIEE296 pKa = 4.05VLIGRR301 pKa = 11.84FLTGNFVPGPHH312 pKa = 6.03STGQPTILVSNNAVTTTYY330 pKa = 10.89SNNGIITQNFGSSGNDD346 pKa = 2.85SLQWQNAAGVIGNVSIDD363 pKa = 3.13ASGAIRR369 pKa = 11.84IDD371 pKa = 3.45ANNATEE377 pKa = 4.09VAVIDD382 pKa = 3.53SDD384 pKa = 4.61SRR386 pKa = 11.84IDD388 pKa = 3.66YY389 pKa = 10.58VKK391 pKa = 10.92GSGARR396 pKa = 11.84IVGSSGDD403 pKa = 3.35QDD405 pKa = 3.68TLYY408 pKa = 11.14GGVGSSTYY416 pKa = 10.87VLQTLTTGIKK426 pKa = 9.38DD427 pKa = 3.85TIDD430 pKa = 3.87DD431 pKa = 3.77QDD433 pKa = 4.08GLGSLYY439 pKa = 10.92VGSNLINGKK448 pKa = 9.12GAQTGQQANSWSDD461 pKa = 3.55ANGTLYY467 pKa = 10.74QFVPLSADD475 pKa = 3.4AQIGTMLISGGNFIGPGQIAINDD498 pKa = 3.65FDD500 pKa = 4.43LGKK503 pKa = 10.58AVTDD507 pKa = 3.98ANGDD511 pKa = 3.89LGIHH515 pKa = 6.84LSQEE519 pKa = 3.65IGLTADD525 pKa = 3.26TSEE528 pKa = 4.47TNLFANGMPANQKK541 pKa = 10.83ADD543 pKa = 3.43VALGNSQTFTVHH555 pKa = 7.11ALADD559 pKa = 3.54STAAQTIQLALSGGGSYY576 pKa = 11.05ALSTGATQTAFNGKK590 pKa = 9.47ISVTIAAGQDD600 pKa = 3.1SATFSLIDD608 pKa = 4.65LSNTNQADD616 pKa = 3.94APTLTATYY624 pKa = 9.26TDD626 pKa = 3.58TNGTVTSNTLAITFDD641 pKa = 4.0HH642 pKa = 7.12PAADD646 pKa = 4.19PSTPPTVVGTVSTAPVSIDD665 pKa = 2.98GSKK668 pKa = 10.94YY669 pKa = 10.72NYY671 pKa = 10.0TEE673 pKa = 3.91FNGDD677 pKa = 3.49GLVTGNSGGNGILTGDD693 pKa = 3.96GNSVINGSGALDD705 pKa = 3.7VIIAGNGNNQIYY717 pKa = 10.9ANVQTDD723 pKa = 3.4VATALTQAKK732 pKa = 9.32AAQASGKK739 pKa = 10.15RR740 pKa = 11.84GDD742 pKa = 4.76LISVGNGNNTIVGGNGNDD760 pKa = 3.36AVFTGSGNNTLILGSGAVTVTGGVIVHH787 pKa = 6.07SPISGEE793 pKa = 4.47GVANWASSTEE803 pKa = 3.85FSSDD807 pKa = 2.98VGAEE811 pKa = 4.09TTDD814 pKa = 2.73TGGIVRR820 pKa = 11.84DD821 pKa = 3.65STGFNAPNPYY831 pKa = 8.04YY832 pKa = 11.03ANYY835 pKa = 10.17ISDD838 pKa = 3.94TPVGIGNDD846 pKa = 3.93TIFGGTGNSVYY857 pKa = 10.89YY858 pKa = 10.65LSNGNNYY865 pKa = 9.98LDD867 pKa = 4.19AGGGNDD873 pKa = 4.62FIGAGVGNNTIFGGIGNDD891 pKa = 3.88TIRR894 pKa = 11.84GAGGNNYY901 pKa = 9.42INAEE905 pKa = 4.15SGNDD909 pKa = 3.93LIAAYY914 pKa = 9.8GGNNTIVGGTGNDD927 pKa = 3.89TIFSGDD933 pKa = 3.41NTSNWASSITTEE945 pKa = 3.73NNYY948 pKa = 10.09INGGSGKK955 pKa = 10.07SVIYY959 pKa = 10.21GAGGNDD965 pKa = 3.53TLIGGTGNVTIYY977 pKa = 10.96GGDD980 pKa = 3.39GTEE983 pKa = 5.26DD984 pKa = 2.93ITGGSGHH991 pKa = 7.04DD992 pKa = 3.45VLYY995 pKa = 10.74GGSGTDD1001 pKa = 3.53VIHH1004 pKa = 7.05AGDD1007 pKa = 4.26GGTADD1012 pKa = 3.88SATSIYY1018 pKa = 10.56AGSGNTTIYY1027 pKa = 10.82GGGGTDD1033 pKa = 3.41VIYY1036 pKa = 10.51GSSGTEE1042 pKa = 3.48VIYY1045 pKa = 11.05AGDD1048 pKa = 3.92GGNATAATQIHH1059 pKa = 6.76AGNGNVTVYY1068 pKa = 10.91GGLGTDD1074 pKa = 3.98QIAGGSGTDD1083 pKa = 3.16VLYY1086 pKa = 11.05AGDD1089 pKa = 4.71GGTADD1094 pKa = 3.7GPTTVFAGTGSDD1106 pKa = 3.29TLYY1109 pKa = 11.1GGAGVDD1115 pKa = 4.92ILDD1118 pKa = 3.86ATNGNNVLLVAGSGDD1133 pKa = 3.39ATLLGGSGDD1142 pKa = 3.8DD1143 pKa = 3.56TLIAGGGTDD1152 pKa = 3.89KK1153 pKa = 11.3LVGGAGNNAYY1163 pKa = 10.06VFNAGFGQAEE1173 pKa = 4.21ISSTSGSDD1181 pKa = 3.43VIQFGAGISASDD1193 pKa = 3.72LTVTAALAADD1203 pKa = 4.25GSGVLEE1209 pKa = 4.1IEE1211 pKa = 4.33GAGTILVDD1219 pKa = 3.75SGLSSAVSAVNFADD1233 pKa = 4.14GTSLTIAQLIAQTGAQSEE1251 pKa = 4.82TIAGATGNILFSTGAGDD1268 pKa = 3.97SVQGGSGNDD1277 pKa = 3.48TLSAWGGKK1285 pKa = 7.94ATLTAGSGNNVLYY1298 pKa = 10.87AGGGNDD1304 pKa = 4.54LLVGAAGNDD1313 pKa = 3.64TLIAGAGNDD1322 pKa = 3.78TLISGTGADD1331 pKa = 3.53TLVSNSSGTTFVVNNVQDD1349 pKa = 4.46VIQLPSSQNQNDD1361 pKa = 4.51TIVSSVDD1368 pKa = 3.27YY1369 pKa = 9.98VLPDD1373 pKa = 5.14AITTLTLTGSANLAGTGNTWNSVITGNAGNDD1404 pKa = 2.84ILTAVSGEE1412 pKa = 4.12TTLIAGAGTDD1422 pKa = 3.68TMIGSAGDD1430 pKa = 3.41TVFVVNSLNDD1440 pKa = 3.4VLVEE1444 pKa = 4.01QAVHH1448 pKa = 5.93GTATVQTNLSYY1459 pKa = 10.74TLASNFDD1466 pKa = 3.75NLILTGSANLVGTGNDD1482 pKa = 3.89LNNQLTANGGNDD1494 pKa = 3.4TLIAGGGNDD1503 pKa = 3.67TLLGGSGNDD1512 pKa = 3.16RR1513 pKa = 11.84LIAGSGNDD1521 pKa = 3.95SLNGGAGIDD1530 pKa = 3.64TLVAGSGNDD1539 pKa = 3.26VMSAQTGSTYY1549 pKa = 10.86VFNQGFGQTEE1559 pKa = 4.44IYY1561 pKa = 10.01NAAGSTLQFGAGIAPSDD1578 pKa = 3.72LTLSLTLYY1586 pKa = 10.69GNTPSIILTDD1596 pKa = 3.76SAGGTVTVDD1605 pKa = 3.46GGLSTANNTTYY1616 pKa = 11.41AFADD1620 pKa = 4.24GATLNLNQLIAQTQSTPVTLAGQEE1644 pKa = 4.23GNLVFAAHH1652 pKa = 6.83GNASLVGSSGNDD1664 pKa = 3.24TLYY1667 pKa = 11.11GWGNSDD1673 pKa = 3.55TLVAGSGNNTLYY1685 pKa = 11.28GEE1687 pKa = 4.62DD1688 pKa = 4.0ANDD1691 pKa = 3.38ILVGGTGLDD1700 pKa = 3.4TLYY1703 pKa = 11.16GGTGNDD1709 pKa = 3.09TMIAGTGEE1717 pKa = 3.91NTLVGGQSNDD1727 pKa = 2.95TFMLTEE1733 pKa = 4.56GGVTTINSSTQSGIEE1748 pKa = 4.09TLWLPEE1754 pKa = 3.91KK1755 pKa = 10.22MRR1757 pKa = 11.84LSDD1760 pKa = 4.05FFAVQVGQDD1769 pKa = 3.48LYY1771 pKa = 10.98INSNSLDD1778 pKa = 3.29TTTIINGYY1786 pKa = 10.19FNTQPQSVGWVLGGDD1801 pKa = 3.35NDD1803 pKa = 4.57SPVFLQTWVAAQQANLTGGAIDD1825 pKa = 3.91YY1826 pKa = 10.19SSKK1829 pKa = 10.48ISALEE1834 pKa = 3.75QAYY1837 pKa = 9.31KK1838 pKa = 10.77AQLNADD1844 pKa = 3.72LTSIGKK1850 pKa = 9.55NGTWLDD1856 pKa = 3.84DD1857 pKa = 3.72SVGRR1861 pKa = 11.84ANVGDD1866 pKa = 4.23YY1867 pKa = 11.03YY1868 pKa = 11.49SNNASGVAGTVVNYY1882 pKa = 9.73LFNGVRR1888 pKa = 11.84LDD1890 pKa = 3.36KK1891 pKa = 10.68LTVSGGNLTLSSIDD1905 pKa = 4.12DD1906 pKa = 4.01NVDD1909 pKa = 3.03TTQVHH1914 pKa = 5.33EE1915 pKa = 4.4TTVTGFKK1922 pKa = 9.53TVPVYY1927 pKa = 10.97KK1928 pKa = 10.41LVQLTTGTVVDD1939 pKa = 4.43MGPPSSTGGGSNLSSDD1955 pKa = 4.52EE1956 pKa = 5.01LNLLPNGDD1964 pKa = 3.36VGFIVPGLMRR1974 pKa = 11.84EE1975 pKa = 4.31VQAGTISQAYY1985 pKa = 7.51TYY1987 pKa = 10.08QASTVDD1993 pKa = 3.41VQRR1996 pKa = 11.84TLNTYY2001 pKa = 10.63AITGDD2006 pKa = 3.5GGNDD2010 pKa = 3.37VIMMVSNDD2018 pKa = 3.56ASNGIQSASFMGTVNTGDD2036 pKa = 3.28GNVYY2040 pKa = 10.55VNLGTTSSINPDD2052 pKa = 3.61FGAGQDD2058 pKa = 3.82YY2059 pKa = 10.86SSPEE2063 pKa = 3.91PSSDD2067 pKa = 2.72RR2068 pKa = 11.84SFIVAGAGNDD2078 pKa = 3.81TLIGTDD2084 pKa = 3.54GADD2087 pKa = 3.64TIVGGSGFDD2096 pKa = 3.61YY2097 pKa = 10.57MDD2099 pKa = 4.53GGLGTNTYY2107 pKa = 8.58YY2108 pKa = 11.43VSMHH2112 pKa = 6.55GDD2114 pKa = 3.11ATDD2117 pKa = 4.54IIDD2120 pKa = 4.4DD2121 pKa = 4.03TGDD2124 pKa = 4.01MEE2126 pKa = 5.22NSWQIQEE2133 pKa = 4.57GYY2135 pKa = 10.69GGVVPNKK2142 pKa = 9.06TLVMPDD2148 pKa = 2.91GLLAKK2153 pKa = 9.91DD2154 pKa = 3.47LNYY2157 pKa = 10.63RR2158 pKa = 11.84IVQDD2162 pKa = 3.4PAYY2165 pKa = 8.86PGSSILQINYY2175 pKa = 7.06GTSNVWVVYY2184 pKa = 9.11QEE2186 pKa = 5.06GILEE2190 pKa = 4.14STMYY2194 pKa = 10.69GSNPAVLSIGVNRR2207 pKa = 11.84IAFSDD2212 pKa = 4.19GSTMTLDD2219 pKa = 3.37QFMAAAHH2226 pKa = 5.72QMADD2230 pKa = 3.75DD2231 pKa = 4.0YY2232 pKa = 10.34TPSVSASDD2240 pKa = 3.89QVLQLNQTVAVSGLFTASDD2259 pKa = 3.55TGSNAITWYY2268 pKa = 9.99RR2269 pKa = 11.84VSNSGIDD2276 pKa = 3.16SGYY2279 pKa = 8.73FTLNGKK2285 pKa = 8.59RR2286 pKa = 11.84QSSDD2290 pKa = 2.79APIYY2294 pKa = 8.44LTKK2297 pKa = 10.21PQLSGLQYY2305 pKa = 9.81VTGATAGDD2313 pKa = 4.2DD2314 pKa = 4.56LIQVSAFDD2322 pKa = 3.52GAAWSVVAPISIVTTSNNVLQATSSNQVLTGTAAAADD2359 pKa = 3.99VLIGGYY2365 pKa = 11.0ANDD2368 pKa = 3.95TLVGGGTQDD2377 pKa = 2.76TFFLRR2382 pKa = 11.84RR2383 pKa = 11.84GDD2385 pKa = 3.75GNVVVSEE2392 pKa = 4.43ASDD2395 pKa = 3.56TNGNNTLRR2403 pKa = 11.84FGGGVTSDD2411 pKa = 4.55DD2412 pKa = 4.46LQLTQQGSDD2421 pKa = 3.29VLVKK2425 pKa = 10.79YY2426 pKa = 7.43GTSSDD2431 pKa = 3.61SVLVKK2436 pKa = 10.87NLDD2439 pKa = 3.95TFSGTATIDD2448 pKa = 3.0KK2449 pKa = 10.11FQFADD2454 pKa = 4.38GSHH2457 pKa = 6.08SAYY2460 pKa = 10.47SSSGQGDD2467 pKa = 3.91FLVTNYY2473 pKa = 10.93DD2474 pKa = 3.3STGNKK2479 pKa = 9.08TGDD2482 pKa = 2.95QWQHH2486 pKa = 4.95TDD2488 pKa = 2.68GTTGFDD2494 pKa = 3.04RR2495 pKa = 11.84VASDD2499 pKa = 3.43GTTEE2503 pKa = 4.17TAVTTTNSDD2512 pKa = 2.68GSTYY2516 pKa = 10.09STDD2519 pKa = 3.19QIAYY2523 pKa = 10.4ANGTSQQSWSRR2534 pKa = 11.84SDD2536 pKa = 3.31GAAGSTNTNTAGVVKK2551 pKa = 10.33GSSSVSINGSQEE2563 pKa = 3.43IDD2565 pKa = 3.39SGSGHH2570 pKa = 6.09VLVGSPGADD2579 pKa = 3.21AISGSDD2585 pKa = 4.4DD2586 pKa = 3.32NALLIGGTGNDD2597 pKa = 4.21TITTGTGGNVIGFNLGDD2614 pKa = 3.89GQDD2617 pKa = 3.41TVVANAGDD2625 pKa = 4.23SNVLSLGGKK2634 pKa = 9.9FSYY2637 pKa = 11.26ADD2639 pKa = 3.85LAFQKK2644 pKa = 10.7NGNNLILDD2652 pKa = 4.02VSSSDD2657 pKa = 5.28AITLQDD2663 pKa = 3.58WYY2665 pKa = 10.92ASSDD2669 pKa = 3.55NQQLVTLQVIEE2680 pKa = 4.69AATADD2685 pKa = 3.75YY2686 pKa = 10.75SSRR2689 pKa = 11.84SIDD2692 pKa = 3.27TLKK2695 pKa = 9.11NTKK2698 pKa = 9.93VEE2700 pKa = 4.24TFDD2703 pKa = 3.61FQQLVEE2709 pKa = 5.1AFDD2712 pKa = 3.7QAQIDD2717 pKa = 4.05NPTRR2721 pKa = 11.84GAWSLSSSLLDD2732 pKa = 3.3AHH2734 pKa = 7.25LSNSDD2739 pKa = 3.15TTALGGDD2746 pKa = 3.84LAYY2749 pKa = 10.24EE2750 pKa = 3.95YY2751 pKa = 10.94GVRR2754 pKa = 11.84GNLTGFNVAAAEE2766 pKa = 4.09TVIANTQFATSPQNLHH2782 pKa = 6.15PWGSVSGTAAQIRR2795 pKa = 3.91
MM1 pKa = 7.87TDD3 pKa = 2.78THH5 pKa = 6.7TPSQMPFDD13 pKa = 4.78AGQLQKK19 pKa = 10.45IRR21 pKa = 11.84TLLKK25 pKa = 10.31QQGLLDD31 pKa = 4.22ANGKK35 pKa = 9.36VVDD38 pKa = 4.07TLATFGNSPPKK49 pKa = 10.53GAFAPIYY56 pKa = 10.72AEE58 pKa = 3.5ISKK61 pKa = 10.29IISAVGSTVDD71 pKa = 4.3PKK73 pKa = 9.95IQYY76 pKa = 8.01WFSKK80 pKa = 10.58AADD83 pKa = 3.54INNDD87 pKa = 3.35VTSDD91 pKa = 3.62PADD94 pKa = 3.88DD95 pKa = 4.35YY96 pKa = 11.72IRR98 pKa = 11.84GVTQYY103 pKa = 11.07GLAMRR108 pKa = 11.84HH109 pKa = 5.9KK110 pKa = 10.86LDD112 pKa = 4.24GLSQSAIDD120 pKa = 4.51IGLQQTSNLIGEE132 pKa = 4.3NVINEE137 pKa = 3.91ILGDD141 pKa = 3.68GGIPDD146 pKa = 4.71FGYY149 pKa = 9.81MISGDD154 pKa = 3.03ISAAIGNDD162 pKa = 2.9ALGRR166 pKa = 11.84FQQDD170 pKa = 2.43AGGWGGSFYY179 pKa = 11.21YY180 pKa = 10.15WNAPLNYY187 pKa = 8.05GTTLPVISVGNVIANGGTVLTNGAAVTYY215 pKa = 9.23PSSLKK220 pKa = 10.14EE221 pKa = 3.55FYY223 pKa = 10.09YY224 pKa = 11.22ANSNASADD232 pKa = 3.65FAIPEE237 pKa = 4.15ISTVAASSSVGGILGTLITQGGLPNFPKK265 pKa = 10.45AVNDD269 pKa = 3.04AWVAVLSGSQAQTPIMVKK287 pKa = 10.58AKK289 pKa = 10.47LFADD293 pKa = 4.66TIEE296 pKa = 4.05VLIGRR301 pKa = 11.84FLTGNFVPGPHH312 pKa = 6.03STGQPTILVSNNAVTTTYY330 pKa = 10.89SNNGIITQNFGSSGNDD346 pKa = 2.85SLQWQNAAGVIGNVSIDD363 pKa = 3.13ASGAIRR369 pKa = 11.84IDD371 pKa = 3.45ANNATEE377 pKa = 4.09VAVIDD382 pKa = 3.53SDD384 pKa = 4.61SRR386 pKa = 11.84IDD388 pKa = 3.66YY389 pKa = 10.58VKK391 pKa = 10.92GSGARR396 pKa = 11.84IVGSSGDD403 pKa = 3.35QDD405 pKa = 3.68TLYY408 pKa = 11.14GGVGSSTYY416 pKa = 10.87VLQTLTTGIKK426 pKa = 9.38DD427 pKa = 3.85TIDD430 pKa = 3.87DD431 pKa = 3.77QDD433 pKa = 4.08GLGSLYY439 pKa = 10.92VGSNLINGKK448 pKa = 9.12GAQTGQQANSWSDD461 pKa = 3.55ANGTLYY467 pKa = 10.74QFVPLSADD475 pKa = 3.4AQIGTMLISGGNFIGPGQIAINDD498 pKa = 3.65FDD500 pKa = 4.43LGKK503 pKa = 10.58AVTDD507 pKa = 3.98ANGDD511 pKa = 3.89LGIHH515 pKa = 6.84LSQEE519 pKa = 3.65IGLTADD525 pKa = 3.26TSEE528 pKa = 4.47TNLFANGMPANQKK541 pKa = 10.83ADD543 pKa = 3.43VALGNSQTFTVHH555 pKa = 7.11ALADD559 pKa = 3.54STAAQTIQLALSGGGSYY576 pKa = 11.05ALSTGATQTAFNGKK590 pKa = 9.47ISVTIAAGQDD600 pKa = 3.1SATFSLIDD608 pKa = 4.65LSNTNQADD616 pKa = 3.94APTLTATYY624 pKa = 9.26TDD626 pKa = 3.58TNGTVTSNTLAITFDD641 pKa = 4.0HH642 pKa = 7.12PAADD646 pKa = 4.19PSTPPTVVGTVSTAPVSIDD665 pKa = 2.98GSKK668 pKa = 10.94YY669 pKa = 10.72NYY671 pKa = 10.0TEE673 pKa = 3.91FNGDD677 pKa = 3.49GLVTGNSGGNGILTGDD693 pKa = 3.96GNSVINGSGALDD705 pKa = 3.7VIIAGNGNNQIYY717 pKa = 10.9ANVQTDD723 pKa = 3.4VATALTQAKK732 pKa = 9.32AAQASGKK739 pKa = 10.15RR740 pKa = 11.84GDD742 pKa = 4.76LISVGNGNNTIVGGNGNDD760 pKa = 3.36AVFTGSGNNTLILGSGAVTVTGGVIVHH787 pKa = 6.07SPISGEE793 pKa = 4.47GVANWASSTEE803 pKa = 3.85FSSDD807 pKa = 2.98VGAEE811 pKa = 4.09TTDD814 pKa = 2.73TGGIVRR820 pKa = 11.84DD821 pKa = 3.65STGFNAPNPYY831 pKa = 8.04YY832 pKa = 11.03ANYY835 pKa = 10.17ISDD838 pKa = 3.94TPVGIGNDD846 pKa = 3.93TIFGGTGNSVYY857 pKa = 10.89YY858 pKa = 10.65LSNGNNYY865 pKa = 9.98LDD867 pKa = 4.19AGGGNDD873 pKa = 4.62FIGAGVGNNTIFGGIGNDD891 pKa = 3.88TIRR894 pKa = 11.84GAGGNNYY901 pKa = 9.42INAEE905 pKa = 4.15SGNDD909 pKa = 3.93LIAAYY914 pKa = 9.8GGNNTIVGGTGNDD927 pKa = 3.89TIFSGDD933 pKa = 3.41NTSNWASSITTEE945 pKa = 3.73NNYY948 pKa = 10.09INGGSGKK955 pKa = 10.07SVIYY959 pKa = 10.21GAGGNDD965 pKa = 3.53TLIGGTGNVTIYY977 pKa = 10.96GGDD980 pKa = 3.39GTEE983 pKa = 5.26DD984 pKa = 2.93ITGGSGHH991 pKa = 7.04DD992 pKa = 3.45VLYY995 pKa = 10.74GGSGTDD1001 pKa = 3.53VIHH1004 pKa = 7.05AGDD1007 pKa = 4.26GGTADD1012 pKa = 3.88SATSIYY1018 pKa = 10.56AGSGNTTIYY1027 pKa = 10.82GGGGTDD1033 pKa = 3.41VIYY1036 pKa = 10.51GSSGTEE1042 pKa = 3.48VIYY1045 pKa = 11.05AGDD1048 pKa = 3.92GGNATAATQIHH1059 pKa = 6.76AGNGNVTVYY1068 pKa = 10.91GGLGTDD1074 pKa = 3.98QIAGGSGTDD1083 pKa = 3.16VLYY1086 pKa = 11.05AGDD1089 pKa = 4.71GGTADD1094 pKa = 3.7GPTTVFAGTGSDD1106 pKa = 3.29TLYY1109 pKa = 11.1GGAGVDD1115 pKa = 4.92ILDD1118 pKa = 3.86ATNGNNVLLVAGSGDD1133 pKa = 3.39ATLLGGSGDD1142 pKa = 3.8DD1143 pKa = 3.56TLIAGGGTDD1152 pKa = 3.89KK1153 pKa = 11.3LVGGAGNNAYY1163 pKa = 10.06VFNAGFGQAEE1173 pKa = 4.21ISSTSGSDD1181 pKa = 3.43VIQFGAGISASDD1193 pKa = 3.72LTVTAALAADD1203 pKa = 4.25GSGVLEE1209 pKa = 4.1IEE1211 pKa = 4.33GAGTILVDD1219 pKa = 3.75SGLSSAVSAVNFADD1233 pKa = 4.14GTSLTIAQLIAQTGAQSEE1251 pKa = 4.82TIAGATGNILFSTGAGDD1268 pKa = 3.97SVQGGSGNDD1277 pKa = 3.48TLSAWGGKK1285 pKa = 7.94ATLTAGSGNNVLYY1298 pKa = 10.87AGGGNDD1304 pKa = 4.54LLVGAAGNDD1313 pKa = 3.64TLIAGAGNDD1322 pKa = 3.78TLISGTGADD1331 pKa = 3.53TLVSNSSGTTFVVNNVQDD1349 pKa = 4.46VIQLPSSQNQNDD1361 pKa = 4.51TIVSSVDD1368 pKa = 3.27YY1369 pKa = 9.98VLPDD1373 pKa = 5.14AITTLTLTGSANLAGTGNTWNSVITGNAGNDD1404 pKa = 2.84ILTAVSGEE1412 pKa = 4.12TTLIAGAGTDD1422 pKa = 3.68TMIGSAGDD1430 pKa = 3.41TVFVVNSLNDD1440 pKa = 3.4VLVEE1444 pKa = 4.01QAVHH1448 pKa = 5.93GTATVQTNLSYY1459 pKa = 10.74TLASNFDD1466 pKa = 3.75NLILTGSANLVGTGNDD1482 pKa = 3.89LNNQLTANGGNDD1494 pKa = 3.4TLIAGGGNDD1503 pKa = 3.67TLLGGSGNDD1512 pKa = 3.16RR1513 pKa = 11.84LIAGSGNDD1521 pKa = 3.95SLNGGAGIDD1530 pKa = 3.64TLVAGSGNDD1539 pKa = 3.26VMSAQTGSTYY1549 pKa = 10.86VFNQGFGQTEE1559 pKa = 4.44IYY1561 pKa = 10.01NAAGSTLQFGAGIAPSDD1578 pKa = 3.72LTLSLTLYY1586 pKa = 10.69GNTPSIILTDD1596 pKa = 3.76SAGGTVTVDD1605 pKa = 3.46GGLSTANNTTYY1616 pKa = 11.41AFADD1620 pKa = 4.24GATLNLNQLIAQTQSTPVTLAGQEE1644 pKa = 4.23GNLVFAAHH1652 pKa = 6.83GNASLVGSSGNDD1664 pKa = 3.24TLYY1667 pKa = 11.11GWGNSDD1673 pKa = 3.55TLVAGSGNNTLYY1685 pKa = 11.28GEE1687 pKa = 4.62DD1688 pKa = 4.0ANDD1691 pKa = 3.38ILVGGTGLDD1700 pKa = 3.4TLYY1703 pKa = 11.16GGTGNDD1709 pKa = 3.09TMIAGTGEE1717 pKa = 3.91NTLVGGQSNDD1727 pKa = 2.95TFMLTEE1733 pKa = 4.56GGVTTINSSTQSGIEE1748 pKa = 4.09TLWLPEE1754 pKa = 3.91KK1755 pKa = 10.22MRR1757 pKa = 11.84LSDD1760 pKa = 4.05FFAVQVGQDD1769 pKa = 3.48LYY1771 pKa = 10.98INSNSLDD1778 pKa = 3.29TTTIINGYY1786 pKa = 10.19FNTQPQSVGWVLGGDD1801 pKa = 3.35NDD1803 pKa = 4.57SPVFLQTWVAAQQANLTGGAIDD1825 pKa = 3.91YY1826 pKa = 10.19SSKK1829 pKa = 10.48ISALEE1834 pKa = 3.75QAYY1837 pKa = 9.31KK1838 pKa = 10.77AQLNADD1844 pKa = 3.72LTSIGKK1850 pKa = 9.55NGTWLDD1856 pKa = 3.84DD1857 pKa = 3.72SVGRR1861 pKa = 11.84ANVGDD1866 pKa = 4.23YY1867 pKa = 11.03YY1868 pKa = 11.49SNNASGVAGTVVNYY1882 pKa = 9.73LFNGVRR1888 pKa = 11.84LDD1890 pKa = 3.36KK1891 pKa = 10.68LTVSGGNLTLSSIDD1905 pKa = 4.12DD1906 pKa = 4.01NVDD1909 pKa = 3.03TTQVHH1914 pKa = 5.33EE1915 pKa = 4.4TTVTGFKK1922 pKa = 9.53TVPVYY1927 pKa = 10.97KK1928 pKa = 10.41LVQLTTGTVVDD1939 pKa = 4.43MGPPSSTGGGSNLSSDD1955 pKa = 4.52EE1956 pKa = 5.01LNLLPNGDD1964 pKa = 3.36VGFIVPGLMRR1974 pKa = 11.84EE1975 pKa = 4.31VQAGTISQAYY1985 pKa = 7.51TYY1987 pKa = 10.08QASTVDD1993 pKa = 3.41VQRR1996 pKa = 11.84TLNTYY2001 pKa = 10.63AITGDD2006 pKa = 3.5GGNDD2010 pKa = 3.37VIMMVSNDD2018 pKa = 3.56ASNGIQSASFMGTVNTGDD2036 pKa = 3.28GNVYY2040 pKa = 10.55VNLGTTSSINPDD2052 pKa = 3.61FGAGQDD2058 pKa = 3.82YY2059 pKa = 10.86SSPEE2063 pKa = 3.91PSSDD2067 pKa = 2.72RR2068 pKa = 11.84SFIVAGAGNDD2078 pKa = 3.81TLIGTDD2084 pKa = 3.54GADD2087 pKa = 3.64TIVGGSGFDD2096 pKa = 3.61YY2097 pKa = 10.57MDD2099 pKa = 4.53GGLGTNTYY2107 pKa = 8.58YY2108 pKa = 11.43VSMHH2112 pKa = 6.55GDD2114 pKa = 3.11ATDD2117 pKa = 4.54IIDD2120 pKa = 4.4DD2121 pKa = 4.03TGDD2124 pKa = 4.01MEE2126 pKa = 5.22NSWQIQEE2133 pKa = 4.57GYY2135 pKa = 10.69GGVVPNKK2142 pKa = 9.06TLVMPDD2148 pKa = 2.91GLLAKK2153 pKa = 9.91DD2154 pKa = 3.47LNYY2157 pKa = 10.63RR2158 pKa = 11.84IVQDD2162 pKa = 3.4PAYY2165 pKa = 8.86PGSSILQINYY2175 pKa = 7.06GTSNVWVVYY2184 pKa = 9.11QEE2186 pKa = 5.06GILEE2190 pKa = 4.14STMYY2194 pKa = 10.69GSNPAVLSIGVNRR2207 pKa = 11.84IAFSDD2212 pKa = 4.19GSTMTLDD2219 pKa = 3.37QFMAAAHH2226 pKa = 5.72QMADD2230 pKa = 3.75DD2231 pKa = 4.0YY2232 pKa = 10.34TPSVSASDD2240 pKa = 3.89QVLQLNQTVAVSGLFTASDD2259 pKa = 3.55TGSNAITWYY2268 pKa = 9.99RR2269 pKa = 11.84VSNSGIDD2276 pKa = 3.16SGYY2279 pKa = 8.73FTLNGKK2285 pKa = 8.59RR2286 pKa = 11.84QSSDD2290 pKa = 2.79APIYY2294 pKa = 8.44LTKK2297 pKa = 10.21PQLSGLQYY2305 pKa = 9.81VTGATAGDD2313 pKa = 4.2DD2314 pKa = 4.56LIQVSAFDD2322 pKa = 3.52GAAWSVVAPISIVTTSNNVLQATSSNQVLTGTAAAADD2359 pKa = 3.99VLIGGYY2365 pKa = 11.0ANDD2368 pKa = 3.95TLVGGGTQDD2377 pKa = 2.76TFFLRR2382 pKa = 11.84RR2383 pKa = 11.84GDD2385 pKa = 3.75GNVVVSEE2392 pKa = 4.43ASDD2395 pKa = 3.56TNGNNTLRR2403 pKa = 11.84FGGGVTSDD2411 pKa = 4.55DD2412 pKa = 4.46LQLTQQGSDD2421 pKa = 3.29VLVKK2425 pKa = 10.79YY2426 pKa = 7.43GTSSDD2431 pKa = 3.61SVLVKK2436 pKa = 10.87NLDD2439 pKa = 3.95TFSGTATIDD2448 pKa = 3.0KK2449 pKa = 10.11FQFADD2454 pKa = 4.38GSHH2457 pKa = 6.08SAYY2460 pKa = 10.47SSSGQGDD2467 pKa = 3.91FLVTNYY2473 pKa = 10.93DD2474 pKa = 3.3STGNKK2479 pKa = 9.08TGDD2482 pKa = 2.95QWQHH2486 pKa = 4.95TDD2488 pKa = 2.68GTTGFDD2494 pKa = 3.04RR2495 pKa = 11.84VASDD2499 pKa = 3.43GTTEE2503 pKa = 4.17TAVTTTNSDD2512 pKa = 2.68GSTYY2516 pKa = 10.09STDD2519 pKa = 3.19QIAYY2523 pKa = 10.4ANGTSQQSWSRR2534 pKa = 11.84SDD2536 pKa = 3.31GAAGSTNTNTAGVVKK2551 pKa = 10.33GSSSVSINGSQEE2563 pKa = 3.43IDD2565 pKa = 3.39SGSGHH2570 pKa = 6.09VLVGSPGADD2579 pKa = 3.21AISGSDD2585 pKa = 4.4DD2586 pKa = 3.32NALLIGGTGNDD2597 pKa = 4.21TITTGTGGNVIGFNLGDD2614 pKa = 3.89GQDD2617 pKa = 3.41TVVANAGDD2625 pKa = 4.23SNVLSLGGKK2634 pKa = 9.9FSYY2637 pKa = 11.26ADD2639 pKa = 3.85LAFQKK2644 pKa = 10.7NGNNLILDD2652 pKa = 4.02VSSSDD2657 pKa = 5.28AITLQDD2663 pKa = 3.58WYY2665 pKa = 10.92ASSDD2669 pKa = 3.55NQQLVTLQVIEE2680 pKa = 4.69AATADD2685 pKa = 3.75YY2686 pKa = 10.75SSRR2689 pKa = 11.84SIDD2692 pKa = 3.27TLKK2695 pKa = 9.11NTKK2698 pKa = 9.93VEE2700 pKa = 4.24TFDD2703 pKa = 3.61FQQLVEE2709 pKa = 5.1AFDD2712 pKa = 3.7QAQIDD2717 pKa = 4.05NPTRR2721 pKa = 11.84GAWSLSSSLLDD2732 pKa = 3.3AHH2734 pKa = 7.25LSNSDD2739 pKa = 3.15TTALGGDD2746 pKa = 3.84LAYY2749 pKa = 10.24EE2750 pKa = 3.95YY2751 pKa = 10.94GVRR2754 pKa = 11.84GNLTGFNVAAAEE2766 pKa = 4.09TVIANTQFATSPQNLHH2782 pKa = 6.15PWGSVSGTAAQIRR2795 pKa = 3.91
Molecular weight: 282.66 kDa
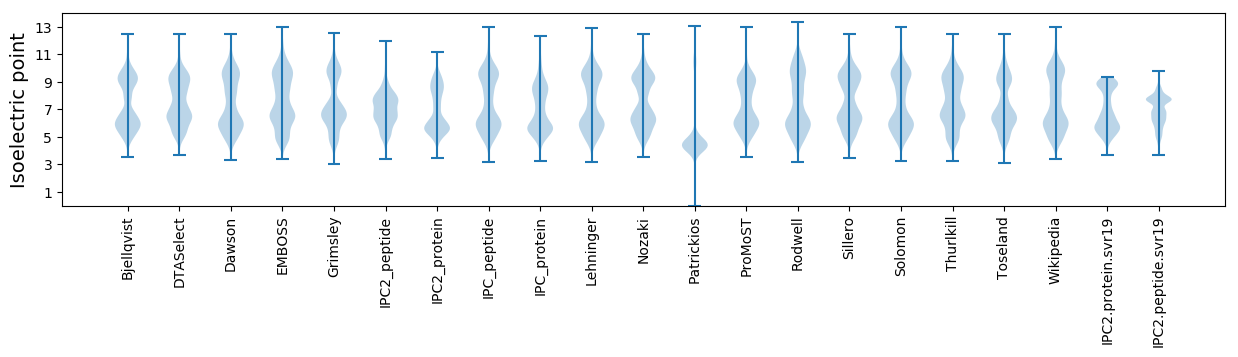
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G0ABE2|G0ABE2_COLFT Putative oxidoreductase FAD/FMN-binding protein OS=Collimonas fungivorans (strain Ter331) OX=1005048 GN=CFU_1516 PE=4 SV=1
MM1 pKa = 7.91PKK3 pKa = 9.63MKK5 pKa = 9.83TKK7 pKa = 10.72SSAKK11 pKa = 9.15KK12 pKa = 9.67RR13 pKa = 11.84FRR15 pKa = 11.84VRR17 pKa = 11.84PGGTVKK23 pKa = 10.62RR24 pKa = 11.84GQAFKK29 pKa = 11.01RR30 pKa = 11.84HH31 pKa = 5.79ILTKK35 pKa = 9.78KK36 pKa = 3.98TTKK39 pKa = 10.23NKK41 pKa = 8.8RR42 pKa = 11.84QLRR45 pKa = 11.84GSVGVHH51 pKa = 4.65EE52 pKa = 4.79TNMASVSRR60 pKa = 11.84MLPTAA65 pKa = 4.04
MM1 pKa = 7.91PKK3 pKa = 9.63MKK5 pKa = 9.83TKK7 pKa = 10.72SSAKK11 pKa = 9.15KK12 pKa = 9.67RR13 pKa = 11.84FRR15 pKa = 11.84VRR17 pKa = 11.84PGGTVKK23 pKa = 10.62RR24 pKa = 11.84GQAFKK29 pKa = 11.01RR30 pKa = 11.84HH31 pKa = 5.79ILTKK35 pKa = 9.78KK36 pKa = 3.98TTKK39 pKa = 10.23NKK41 pKa = 8.8RR42 pKa = 11.84QLRR45 pKa = 11.84GSVGVHH51 pKa = 4.65EE52 pKa = 4.79TNMASVSRR60 pKa = 11.84MLPTAA65 pKa = 4.04
Molecular weight: 7.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1527625 |
30 |
5651 |
344.9 |
37.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.84 ± 0.049 | 0.912 ± 0.012 |
5.173 ± 0.028 | 4.863 ± 0.038 |
3.758 ± 0.026 | 8.05 ± 0.048 |
2.201 ± 0.023 | 5.263 ± 0.026 |
3.961 ± 0.037 | 10.556 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.5 ± 0.018 | 3.361 ± 0.039 |
4.814 ± 0.03 | 4.427 ± 0.029 |
5.881 ± 0.042 | 6.222 ± 0.084 |
5.295 ± 0.06 | 7.064 ± 0.031 |
1.299 ± 0.013 | 2.561 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
