
Macleaya cordata
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Ranunculales; Papaveraceae; Papaveroideae; Macleaya
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
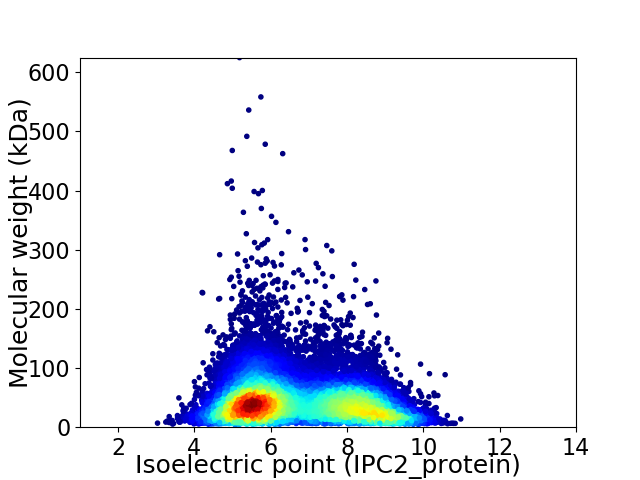
Virtual 2D-PAGE plot for 21697 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A200Q3G1|A0A200Q3G1_9MAGN Cytochrome P450 OS=Macleaya cordata OX=56857 GN=BVC80_1211g57 PE=3 SV=1
MM1 pKa = 7.59EE2 pKa = 5.76NIKK5 pKa = 10.65DD6 pKa = 3.88GPGGAKK12 pKa = 9.89TNDD15 pKa = 3.75EE16 pKa = 4.46NPNSPSEE23 pKa = 4.25AEE25 pKa = 4.39TEE27 pKa = 4.33VEE29 pKa = 4.21TNLASSQQQSSRR41 pKa = 11.84TPFTNLSQIDD51 pKa = 3.81ADD53 pKa = 3.97LALARR58 pKa = 11.84TLQEE62 pKa = 3.58QEE64 pKa = 3.59RR65 pKa = 11.84AYY67 pKa = 11.04MMLRR71 pKa = 11.84MNGEE75 pKa = 3.75MSDD78 pKa = 3.84YY79 pKa = 10.85GSSDD83 pKa = 3.36SGSYY87 pKa = 10.3AYY89 pKa = 10.1EE90 pKa = 5.04DD91 pKa = 5.48DD92 pKa = 5.5VDD94 pKa = 6.32DD95 pKa = 7.03DD96 pKa = 5.65DD97 pKa = 7.39DD98 pKa = 5.14NDD100 pKa = 3.56NHH102 pKa = 6.4NVNNDD107 pKa = 3.32EE108 pKa = 4.01NSEE111 pKa = 3.94IDD113 pKa = 3.88GDD115 pKa = 3.93EE116 pKa = 3.8EE117 pKa = 4.92DD118 pKa = 4.59AFDD121 pKa = 3.87VHH123 pKa = 7.38AHH125 pKa = 6.48ADD127 pKa = 3.46EE128 pKa = 5.38DD129 pKa = 4.23NNQGAEE135 pKa = 4.38LDD137 pKa = 3.79PSAYY141 pKa = 10.57SSDD144 pKa = 3.4EE145 pKa = 3.93AYY147 pKa = 10.61ARR149 pKa = 11.84ALQEE153 pKa = 3.97AEE155 pKa = 4.08EE156 pKa = 5.1RR157 pKa = 11.84EE158 pKa = 3.9MAMRR162 pKa = 11.84LMALAGINDD171 pKa = 4.03FAVEE175 pKa = 4.02AEE177 pKa = 4.6DD178 pKa = 5.72NVDD181 pKa = 3.59HH182 pKa = 7.69DD183 pKa = 5.21SNSQVSYY190 pKa = 11.53EE191 pKa = 4.05EE192 pKa = 4.71LDD194 pKa = 3.73PDD196 pKa = 3.62EE197 pKa = 5.34LSYY200 pKa = 11.76EE201 pKa = 4.06EE202 pKa = 5.32LLALGEE208 pKa = 4.44VVGTEE213 pKa = 4.03SRR215 pKa = 11.84GLSSDD220 pKa = 4.16TIAALPSISYY230 pKa = 9.75KK231 pKa = 10.9AQDD234 pKa = 4.24DD235 pKa = 3.96QDD237 pKa = 4.02GSTDD241 pKa = 3.07QCVICRR247 pKa = 11.84LEE249 pKa = 4.21YY250 pKa = 10.83DD251 pKa = 3.8DD252 pKa = 6.65GEE254 pKa = 4.57TLTMLSCKK262 pKa = 10.0HH263 pKa = 6.45SYY265 pKa = 10.38HH266 pKa = 7.05PEE268 pKa = 4.54CINNWLQINKK278 pKa = 8.6VCPVCSTEE286 pKa = 3.92VSTSEE291 pKa = 4.3SSRR294 pKa = 3.48
MM1 pKa = 7.59EE2 pKa = 5.76NIKK5 pKa = 10.65DD6 pKa = 3.88GPGGAKK12 pKa = 9.89TNDD15 pKa = 3.75EE16 pKa = 4.46NPNSPSEE23 pKa = 4.25AEE25 pKa = 4.39TEE27 pKa = 4.33VEE29 pKa = 4.21TNLASSQQQSSRR41 pKa = 11.84TPFTNLSQIDD51 pKa = 3.81ADD53 pKa = 3.97LALARR58 pKa = 11.84TLQEE62 pKa = 3.58QEE64 pKa = 3.59RR65 pKa = 11.84AYY67 pKa = 11.04MMLRR71 pKa = 11.84MNGEE75 pKa = 3.75MSDD78 pKa = 3.84YY79 pKa = 10.85GSSDD83 pKa = 3.36SGSYY87 pKa = 10.3AYY89 pKa = 10.1EE90 pKa = 5.04DD91 pKa = 5.48DD92 pKa = 5.5VDD94 pKa = 6.32DD95 pKa = 7.03DD96 pKa = 5.65DD97 pKa = 7.39DD98 pKa = 5.14NDD100 pKa = 3.56NHH102 pKa = 6.4NVNNDD107 pKa = 3.32EE108 pKa = 4.01NSEE111 pKa = 3.94IDD113 pKa = 3.88GDD115 pKa = 3.93EE116 pKa = 3.8EE117 pKa = 4.92DD118 pKa = 4.59AFDD121 pKa = 3.87VHH123 pKa = 7.38AHH125 pKa = 6.48ADD127 pKa = 3.46EE128 pKa = 5.38DD129 pKa = 4.23NNQGAEE135 pKa = 4.38LDD137 pKa = 3.79PSAYY141 pKa = 10.57SSDD144 pKa = 3.4EE145 pKa = 3.93AYY147 pKa = 10.61ARR149 pKa = 11.84ALQEE153 pKa = 3.97AEE155 pKa = 4.08EE156 pKa = 5.1RR157 pKa = 11.84EE158 pKa = 3.9MAMRR162 pKa = 11.84LMALAGINDD171 pKa = 4.03FAVEE175 pKa = 4.02AEE177 pKa = 4.6DD178 pKa = 5.72NVDD181 pKa = 3.59HH182 pKa = 7.69DD183 pKa = 5.21SNSQVSYY190 pKa = 11.53EE191 pKa = 4.05EE192 pKa = 4.71LDD194 pKa = 3.73PDD196 pKa = 3.62EE197 pKa = 5.34LSYY200 pKa = 11.76EE201 pKa = 4.06EE202 pKa = 5.32LLALGEE208 pKa = 4.44VVGTEE213 pKa = 4.03SRR215 pKa = 11.84GLSSDD220 pKa = 4.16TIAALPSISYY230 pKa = 9.75KK231 pKa = 10.9AQDD234 pKa = 4.24DD235 pKa = 3.96QDD237 pKa = 4.02GSTDD241 pKa = 3.07QCVICRR247 pKa = 11.84LEE249 pKa = 4.21YY250 pKa = 10.83DD251 pKa = 3.8DD252 pKa = 6.65GEE254 pKa = 4.57TLTMLSCKK262 pKa = 10.0HH263 pKa = 6.45SYY265 pKa = 10.38HH266 pKa = 7.05PEE268 pKa = 4.54CINNWLQINKK278 pKa = 8.6VCPVCSTEE286 pKa = 3.92VSTSEE291 pKa = 4.3SSRR294 pKa = 3.48
Molecular weight: 32.38 kDa
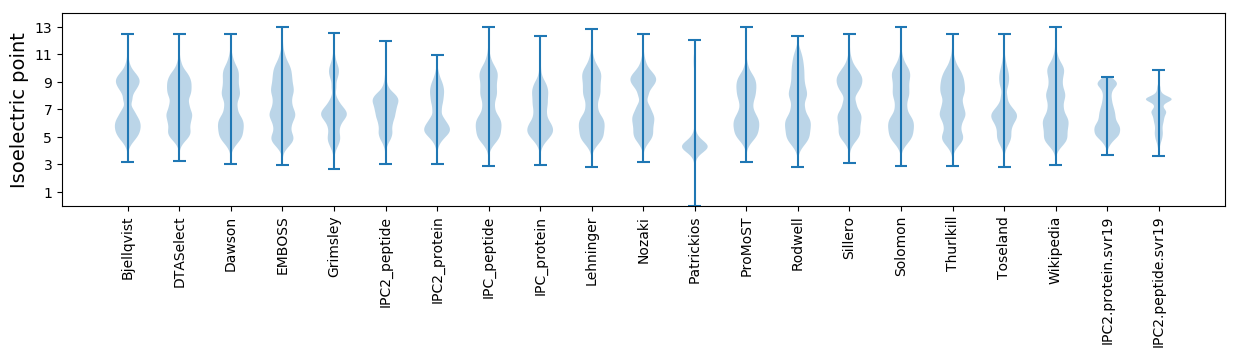
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A200QLK3|A0A200QLK3_9MAGN DNA-binding WRKY OS=Macleaya cordata OX=56857 GN=BVC80_9007g24 PE=4 SV=1
MM1 pKa = 7.71ASHH4 pKa = 6.87SPLNVSNISNLVPIKK19 pKa = 9.84LTRR22 pKa = 11.84EE23 pKa = 3.76NYY25 pKa = 9.84ILWKK29 pKa = 10.62SVFLPILRR37 pKa = 11.84SYY39 pKa = 11.34NLLRR43 pKa = 11.84FVDD46 pKa = 5.23GSHH49 pKa = 7.26PCPSRR54 pKa = 11.84LVTDD58 pKa = 4.19AAGKK62 pKa = 8.08DD63 pKa = 3.71TQATNPDD70 pKa = 3.28FTEE73 pKa = 3.67WRR75 pKa = 11.84QLLTAEE81 pKa = 4.7APPTAFSASTGPRR94 pKa = 11.84TFTRR98 pKa = 11.84APSNQQSTARR108 pKa = 11.84FSKK111 pKa = 10.33DD112 pKa = 2.57KK113 pKa = 10.75HH114 pKa = 5.39KK115 pKa = 11.31ARR117 pKa = 11.84VFYY120 pKa = 10.51RR121 pKa = 11.84GSSSNSGSGSGSGSGSSPRR140 pKa = 11.84TSDD143 pKa = 3.81RR144 pKa = 11.84PVCQICNRR152 pKa = 11.84VGHH155 pKa = 5.56TAIDD159 pKa = 3.36CHH161 pKa = 7.33NRR163 pKa = 11.84MNHH166 pKa = 5.94AFQGRR171 pKa = 11.84IPTQRR176 pKa = 11.84LRR178 pKa = 11.84AMVATNAIHH187 pKa = 6.84ARR189 pKa = 11.84PSLGTEE195 pKa = 4.15GLVTLPLPRR204 pKa = 11.84SSCYY208 pKa = 10.5EE209 pKa = 4.04LVTKK213 pKa = 10.67
MM1 pKa = 7.71ASHH4 pKa = 6.87SPLNVSNISNLVPIKK19 pKa = 9.84LTRR22 pKa = 11.84EE23 pKa = 3.76NYY25 pKa = 9.84ILWKK29 pKa = 10.62SVFLPILRR37 pKa = 11.84SYY39 pKa = 11.34NLLRR43 pKa = 11.84FVDD46 pKa = 5.23GSHH49 pKa = 7.26PCPSRR54 pKa = 11.84LVTDD58 pKa = 4.19AAGKK62 pKa = 8.08DD63 pKa = 3.71TQATNPDD70 pKa = 3.28FTEE73 pKa = 3.67WRR75 pKa = 11.84QLLTAEE81 pKa = 4.7APPTAFSASTGPRR94 pKa = 11.84TFTRR98 pKa = 11.84APSNQQSTARR108 pKa = 11.84FSKK111 pKa = 10.33DD112 pKa = 2.57KK113 pKa = 10.75HH114 pKa = 5.39KK115 pKa = 11.31ARR117 pKa = 11.84VFYY120 pKa = 10.51RR121 pKa = 11.84GSSSNSGSGSGSGSGSSPRR140 pKa = 11.84TSDD143 pKa = 3.81RR144 pKa = 11.84PVCQICNRR152 pKa = 11.84VGHH155 pKa = 5.56TAIDD159 pKa = 3.36CHH161 pKa = 7.33NRR163 pKa = 11.84MNHH166 pKa = 5.94AFQGRR171 pKa = 11.84IPTQRR176 pKa = 11.84LRR178 pKa = 11.84AMVATNAIHH187 pKa = 6.84ARR189 pKa = 11.84PSLGTEE195 pKa = 4.15GLVTLPLPRR204 pKa = 11.84SSCYY208 pKa = 10.5EE209 pKa = 4.04LVTKK213 pKa = 10.67
Molecular weight: 23.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9115806 |
34 |
5524 |
420.1 |
46.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.386 ± 0.015 | 1.812 ± 0.008 |
5.287 ± 0.012 | 6.576 ± 0.018 |
4.23 ± 0.011 | 6.616 ± 0.014 |
2.344 ± 0.006 | 5.479 ± 0.013 |
6.09 ± 0.015 | 9.622 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.45 ± 0.007 | 4.494 ± 0.01 |
4.9 ± 0.016 | 3.683 ± 0.014 |
5.327 ± 0.014 | 9.088 ± 0.02 |
4.984 ± 0.01 | 6.539 ± 0.011 |
1.332 ± 0.006 | 2.758 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
