
Thecamonas trahens ATCC 50062
Taxonomy: cellular organisms; Eukaryota; Apusozoa; Apusomonadidae; Thecamonas; Thecamonas trahens
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
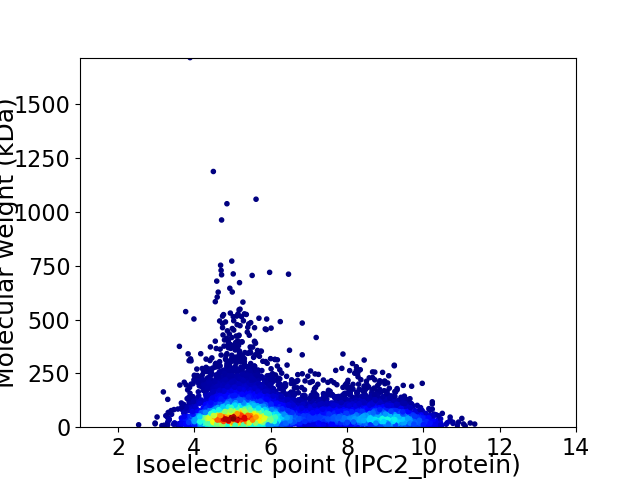
Virtual 2D-PAGE plot for 10604 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0L0DW12|A0A0L0DW12_THETB DnaJ-domain-containing protein 1 OS=Thecamonas trahens ATCC 50062 OX=461836 GN=AMSG_02332 PE=3 SV=1
MM1 pKa = 7.61AVYY4 pKa = 9.89QVLTTMAMVCMLAHH18 pKa = 7.71LARR21 pKa = 11.84GDD23 pKa = 3.49GTTQLGTTQALQASTVLYY41 pKa = 10.92LDD43 pKa = 3.97IDD45 pKa = 4.16NIANQRR51 pKa = 11.84IYY53 pKa = 10.85FQGTGQLRR61 pKa = 11.84IYY63 pKa = 10.45RR64 pKa = 11.84PGDD67 pKa = 3.15FAQVGTILSGQLYY80 pKa = 7.89TPTMTGVYY88 pKa = 9.93GALLTLDD95 pKa = 3.44QTYY98 pKa = 10.3GAVWDD103 pKa = 3.59ISVYY107 pKa = 9.58DD108 pKa = 4.06TSTSSIDD115 pKa = 3.09AGRR118 pKa = 11.84VFSFSWNLASNSFSQSDD135 pKa = 3.8AVDD138 pKa = 3.42VSIWSLFSTSLGAKK152 pKa = 8.98LVEE155 pKa = 4.04NRR157 pKa = 11.84FEE159 pKa = 4.12GMAGFIYY166 pKa = 10.2QIAASTTGIEE176 pKa = 4.27TATPKK181 pKa = 10.56YY182 pKa = 10.51KK183 pKa = 10.5SVLQSGNSVTPNIRR197 pKa = 11.84LYY199 pKa = 10.66LSKK202 pKa = 10.36PEE204 pKa = 4.17HH205 pKa = 5.67VTLSSSVPNIVFEE218 pKa = 4.15GVEE221 pKa = 4.83FVTLGCFDD229 pKa = 5.89FISDD233 pKa = 4.32CNTVVADD240 pKa = 3.98KK241 pKa = 10.27VHH243 pKa = 6.53PRR245 pKa = 11.84FKK247 pKa = 10.86FNTDD251 pKa = 3.02AAGGRR256 pKa = 11.84YY257 pKa = 7.4TLVCDD262 pKa = 4.09TNSDD266 pKa = 3.51GALDD270 pKa = 4.46LVRR273 pKa = 11.84QTDD276 pKa = 3.3TSLRR280 pKa = 11.84GTTVAGDD287 pKa = 4.71NIVEE291 pKa = 3.94WDD293 pKa = 3.65GRR295 pKa = 11.84DD296 pKa = 3.56DD297 pKa = 4.17NNALVTAGFYY307 pKa = 7.36TCKK310 pKa = 9.84MFVYY314 pKa = 10.11LDD316 pKa = 4.45EE317 pKa = 4.21IHH319 pKa = 6.75FPLVDD324 pKa = 3.7MEE326 pKa = 4.51TVYY329 pKa = 10.86PGLRR333 pKa = 11.84MFDD336 pKa = 3.67HH337 pKa = 7.3SSGTPRR343 pKa = 11.84PQIMYY348 pKa = 9.88WNDD351 pKa = 3.33DD352 pKa = 3.61GVNAPIAMPNGQFGATYY369 pKa = 10.07SGCCGIDD376 pKa = 3.13SGVSTDD382 pKa = 4.35PISPNVNARR391 pKa = 11.84SWGNFVSGSKK401 pKa = 10.91GDD403 pKa = 3.7TAILNTFSLVEE414 pKa = 4.18VTVSEE419 pKa = 4.88AINISVAQPSQCEE432 pKa = 3.92TATVNLIDD440 pKa = 3.97YY441 pKa = 9.86VLTEE445 pKa = 4.74LDD447 pKa = 3.14SGTTVVEE454 pKa = 4.15VKK456 pKa = 10.56LAVSSYY462 pKa = 10.41LAGDD466 pKa = 3.08ITLAYY471 pKa = 6.2TTADD475 pKa = 3.34GTATAGSGDD484 pKa = 3.85YY485 pKa = 10.61VATSGSIVISSGDD498 pKa = 3.41FCNSLSVTINGDD510 pKa = 3.74DD511 pKa = 3.13IVEE514 pKa = 3.92YY515 pKa = 10.88DD516 pKa = 3.05EE517 pKa = 5.28WFFLDD522 pKa = 3.55VTQTAGPTVIIADD535 pKa = 3.3GRR537 pKa = 11.84ANVTILNDD545 pKa = 3.71DD546 pKa = 3.96TPVTISMTNSATSVTEE562 pKa = 3.97PSSGSANAGLTFTLSAPSTAPVTVYY587 pKa = 10.81LATTAPGGSLVTSGTDD603 pKa = 3.52FTPPPASIVIPAGVTTFTYY622 pKa = 9.99DD623 pKa = 2.75IPVAADD629 pKa = 3.79RR630 pKa = 11.84FWDD633 pKa = 4.12DD634 pKa = 3.81GEE636 pKa = 4.26QFTVSIAGTSVEE648 pKa = 4.31GAVGGQTSTTFTINDD663 pKa = 3.73SGPAPVVTASVVSGNVASSTGDD685 pKa = 3.15GTASLSSTLPGDD697 pKa = 3.68GRR699 pKa = 11.84VGEE702 pKa = 4.42SNEE705 pKa = 3.78VVYY708 pKa = 10.71QLTLSRR714 pKa = 11.84PSEE717 pKa = 4.11YY718 pKa = 10.77AAGLTLNVDD727 pKa = 4.38GPATTATLPGDD738 pKa = 3.87ADD740 pKa = 3.98LVGTIDD746 pKa = 3.42RR747 pKa = 11.84TGGPFTIAAGEE758 pKa = 4.12TSFNVSLVVAGDD770 pKa = 3.63GDD772 pKa = 4.04AEE774 pKa = 4.26ADD776 pKa = 3.63EE777 pKa = 5.33VVDD780 pKa = 5.12FDD782 pKa = 5.76AVVNLNGASNPTTTATILTNIPNDD806 pKa = 3.72DD807 pKa = 3.69VAVVQMAAPTAATTEE822 pKa = 4.41PGSGTVNVPITVNLLATSPSPVTINIQTSVSSGSLATGGADD863 pKa = 3.49FTAEE867 pKa = 4.06STTVVVPPGQMSVVYY882 pKa = 9.52NAVVAADD889 pKa = 3.22TWYY892 pKa = 10.51DD893 pKa = 3.23GGEE896 pKa = 4.15TFDD899 pKa = 4.23VKK901 pKa = 10.25ITGADD906 pKa = 2.85NGFAVGAADD915 pKa = 3.69TTVVTIADD923 pKa = 3.82SGPAPVLTATVVGGTTTAVSGDD945 pKa = 3.66GTDD948 pKa = 3.89VIDD951 pKa = 3.54VTGPEE956 pKa = 4.44GAASTTNEE964 pKa = 4.13LVVEE968 pKa = 4.6LALSRR973 pKa = 11.84PSEE976 pKa = 4.14YY977 pKa = 10.79AAGLTVQVDD986 pKa = 4.6GVTSTADD993 pKa = 3.35VPSDD997 pKa = 3.24ATIVDD1002 pKa = 4.03TVARR1006 pKa = 11.84TGSPMTIPALATTHH1020 pKa = 5.12QVSVVVSGDD1029 pKa = 3.27GVLEE1033 pKa = 4.02PTEE1036 pKa = 4.16VLDD1039 pKa = 5.31LGLIANYY1046 pKa = 10.3GGAEE1050 pKa = 3.96LAADD1054 pKa = 4.07VVSVTATITNDD1065 pKa = 3.12DD1066 pKa = 3.91AAVVTMAAGTAAATEE1081 pKa = 4.5PGSGTVNVPITQALRR1096 pKa = 3.48
MM1 pKa = 7.61AVYY4 pKa = 9.89QVLTTMAMVCMLAHH18 pKa = 7.71LARR21 pKa = 11.84GDD23 pKa = 3.49GTTQLGTTQALQASTVLYY41 pKa = 10.92LDD43 pKa = 3.97IDD45 pKa = 4.16NIANQRR51 pKa = 11.84IYY53 pKa = 10.85FQGTGQLRR61 pKa = 11.84IYY63 pKa = 10.45RR64 pKa = 11.84PGDD67 pKa = 3.15FAQVGTILSGQLYY80 pKa = 7.89TPTMTGVYY88 pKa = 9.93GALLTLDD95 pKa = 3.44QTYY98 pKa = 10.3GAVWDD103 pKa = 3.59ISVYY107 pKa = 9.58DD108 pKa = 4.06TSTSSIDD115 pKa = 3.09AGRR118 pKa = 11.84VFSFSWNLASNSFSQSDD135 pKa = 3.8AVDD138 pKa = 3.42VSIWSLFSTSLGAKK152 pKa = 8.98LVEE155 pKa = 4.04NRR157 pKa = 11.84FEE159 pKa = 4.12GMAGFIYY166 pKa = 10.2QIAASTTGIEE176 pKa = 4.27TATPKK181 pKa = 10.56YY182 pKa = 10.51KK183 pKa = 10.5SVLQSGNSVTPNIRR197 pKa = 11.84LYY199 pKa = 10.66LSKK202 pKa = 10.36PEE204 pKa = 4.17HH205 pKa = 5.67VTLSSSVPNIVFEE218 pKa = 4.15GVEE221 pKa = 4.83FVTLGCFDD229 pKa = 5.89FISDD233 pKa = 4.32CNTVVADD240 pKa = 3.98KK241 pKa = 10.27VHH243 pKa = 6.53PRR245 pKa = 11.84FKK247 pKa = 10.86FNTDD251 pKa = 3.02AAGGRR256 pKa = 11.84YY257 pKa = 7.4TLVCDD262 pKa = 4.09TNSDD266 pKa = 3.51GALDD270 pKa = 4.46LVRR273 pKa = 11.84QTDD276 pKa = 3.3TSLRR280 pKa = 11.84GTTVAGDD287 pKa = 4.71NIVEE291 pKa = 3.94WDD293 pKa = 3.65GRR295 pKa = 11.84DD296 pKa = 3.56DD297 pKa = 4.17NNALVTAGFYY307 pKa = 7.36TCKK310 pKa = 9.84MFVYY314 pKa = 10.11LDD316 pKa = 4.45EE317 pKa = 4.21IHH319 pKa = 6.75FPLVDD324 pKa = 3.7MEE326 pKa = 4.51TVYY329 pKa = 10.86PGLRR333 pKa = 11.84MFDD336 pKa = 3.67HH337 pKa = 7.3SSGTPRR343 pKa = 11.84PQIMYY348 pKa = 9.88WNDD351 pKa = 3.33DD352 pKa = 3.61GVNAPIAMPNGQFGATYY369 pKa = 10.07SGCCGIDD376 pKa = 3.13SGVSTDD382 pKa = 4.35PISPNVNARR391 pKa = 11.84SWGNFVSGSKK401 pKa = 10.91GDD403 pKa = 3.7TAILNTFSLVEE414 pKa = 4.18VTVSEE419 pKa = 4.88AINISVAQPSQCEE432 pKa = 3.92TATVNLIDD440 pKa = 3.97YY441 pKa = 9.86VLTEE445 pKa = 4.74LDD447 pKa = 3.14SGTTVVEE454 pKa = 4.15VKK456 pKa = 10.56LAVSSYY462 pKa = 10.41LAGDD466 pKa = 3.08ITLAYY471 pKa = 6.2TTADD475 pKa = 3.34GTATAGSGDD484 pKa = 3.85YY485 pKa = 10.61VATSGSIVISSGDD498 pKa = 3.41FCNSLSVTINGDD510 pKa = 3.74DD511 pKa = 3.13IVEE514 pKa = 3.92YY515 pKa = 10.88DD516 pKa = 3.05EE517 pKa = 5.28WFFLDD522 pKa = 3.55VTQTAGPTVIIADD535 pKa = 3.3GRR537 pKa = 11.84ANVTILNDD545 pKa = 3.71DD546 pKa = 3.96TPVTISMTNSATSVTEE562 pKa = 3.97PSSGSANAGLTFTLSAPSTAPVTVYY587 pKa = 10.81LATTAPGGSLVTSGTDD603 pKa = 3.52FTPPPASIVIPAGVTTFTYY622 pKa = 9.99DD623 pKa = 2.75IPVAADD629 pKa = 3.79RR630 pKa = 11.84FWDD633 pKa = 4.12DD634 pKa = 3.81GEE636 pKa = 4.26QFTVSIAGTSVEE648 pKa = 4.31GAVGGQTSTTFTINDD663 pKa = 3.73SGPAPVVTASVVSGNVASSTGDD685 pKa = 3.15GTASLSSTLPGDD697 pKa = 3.68GRR699 pKa = 11.84VGEE702 pKa = 4.42SNEE705 pKa = 3.78VVYY708 pKa = 10.71QLTLSRR714 pKa = 11.84PSEE717 pKa = 4.11YY718 pKa = 10.77AAGLTLNVDD727 pKa = 4.38GPATTATLPGDD738 pKa = 3.87ADD740 pKa = 3.98LVGTIDD746 pKa = 3.42RR747 pKa = 11.84TGGPFTIAAGEE758 pKa = 4.12TSFNVSLVVAGDD770 pKa = 3.63GDD772 pKa = 4.04AEE774 pKa = 4.26ADD776 pKa = 3.63EE777 pKa = 5.33VVDD780 pKa = 5.12FDD782 pKa = 5.76AVVNLNGASNPTTTATILTNIPNDD806 pKa = 3.72DD807 pKa = 3.69VAVVQMAAPTAATTEE822 pKa = 4.41PGSGTVNVPITVNLLATSPSPVTINIQTSVSSGSLATGGADD863 pKa = 3.49FTAEE867 pKa = 4.06STTVVVPPGQMSVVYY882 pKa = 9.52NAVVAADD889 pKa = 3.22TWYY892 pKa = 10.51DD893 pKa = 3.23GGEE896 pKa = 4.15TFDD899 pKa = 4.23VKK901 pKa = 10.25ITGADD906 pKa = 2.85NGFAVGAADD915 pKa = 3.69TTVVTIADD923 pKa = 3.82SGPAPVLTATVVGGTTTAVSGDD945 pKa = 3.66GTDD948 pKa = 3.89VIDD951 pKa = 3.54VTGPEE956 pKa = 4.44GAASTTNEE964 pKa = 4.13LVVEE968 pKa = 4.6LALSRR973 pKa = 11.84PSEE976 pKa = 4.14YY977 pKa = 10.79AAGLTVQVDD986 pKa = 4.6GVTSTADD993 pKa = 3.35VPSDD997 pKa = 3.24ATIVDD1002 pKa = 4.03TVARR1006 pKa = 11.84TGSPMTIPALATTHH1020 pKa = 5.12QVSVVVSGDD1029 pKa = 3.27GVLEE1033 pKa = 4.02PTEE1036 pKa = 4.16VLDD1039 pKa = 5.31LGLIANYY1046 pKa = 10.3GGAEE1050 pKa = 3.96LAADD1054 pKa = 4.07VVSVTATITNDD1065 pKa = 3.12DD1066 pKa = 3.91AAVVTMAAGTAAATEE1081 pKa = 4.5PGSGTVNVPITQALRR1096 pKa = 3.48
Molecular weight: 112.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L0DL00|A0A0L0DL00_THETB Lactamase_B domain-containing protein OS=Thecamonas trahens ATCC 50062 OX=461836 GN=AMSG_11629 PE=4 SV=1
MM1 pKa = 6.61PTARR5 pKa = 11.84ALSTRR10 pKa = 11.84PSSGSSSATPSRR22 pKa = 11.84PASRR26 pKa = 11.84SRR28 pKa = 11.84TRR30 pKa = 11.84IRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84QKK38 pKa = 9.8RR39 pKa = 11.84ARR41 pKa = 11.84ARR43 pKa = 11.84ARR45 pKa = 11.84EE46 pKa = 4.03PARR49 pKa = 11.84AQARR53 pKa = 11.84RR54 pKa = 11.84QRR56 pKa = 11.84HH57 pKa = 3.55RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84KK63 pKa = 6.37QTRR66 pKa = 11.84HH67 pKa = 4.32RR68 pKa = 11.84TQPQPRR74 pKa = 11.84LRR76 pKa = 11.84ARR78 pKa = 11.84ARR80 pKa = 11.84PVARR84 pKa = 11.84TTSIAALRR92 pKa = 11.84PGRR95 pKa = 11.84SWPTRR100 pKa = 11.84RR101 pKa = 11.84GWKK104 pKa = 9.87RR105 pKa = 11.84PVTRR109 pKa = 11.84RR110 pKa = 11.84SAGMTRR116 pKa = 11.84RR117 pKa = 11.84KK118 pKa = 9.85GRR120 pKa = 11.84RR121 pKa = 11.84ADD123 pKa = 3.18AA124 pKa = 4.43
MM1 pKa = 6.61PTARR5 pKa = 11.84ALSTRR10 pKa = 11.84PSSGSSSATPSRR22 pKa = 11.84PASRR26 pKa = 11.84SRR28 pKa = 11.84TRR30 pKa = 11.84IRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84QKK38 pKa = 9.8RR39 pKa = 11.84ARR41 pKa = 11.84ARR43 pKa = 11.84ARR45 pKa = 11.84EE46 pKa = 4.03PARR49 pKa = 11.84AQARR53 pKa = 11.84RR54 pKa = 11.84QRR56 pKa = 11.84HH57 pKa = 3.55RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84KK63 pKa = 6.37QTRR66 pKa = 11.84HH67 pKa = 4.32RR68 pKa = 11.84TQPQPRR74 pKa = 11.84LRR76 pKa = 11.84ARR78 pKa = 11.84ARR80 pKa = 11.84PVARR84 pKa = 11.84TTSIAALRR92 pKa = 11.84PGRR95 pKa = 11.84SWPTRR100 pKa = 11.84RR101 pKa = 11.84GWKK104 pKa = 9.87RR105 pKa = 11.84PVTRR109 pKa = 11.84RR110 pKa = 11.84SAGMTRR116 pKa = 11.84RR117 pKa = 11.84KK118 pKa = 9.85GRR120 pKa = 11.84RR121 pKa = 11.84ADD123 pKa = 3.18AA124 pKa = 4.43
Molecular weight: 14.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6918374 |
49 |
16619 |
652.4 |
69.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.527 ± 0.05 | 1.201 ± 0.014 |
5.752 ± 0.017 | 5.574 ± 0.036 |
3.012 ± 0.017 | 7.239 ± 0.032 |
2.153 ± 0.011 | 3.264 ± 0.015 |
3.295 ± 0.025 | 9.859 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.042 ± 0.01 | 2.349 ± 0.012 |
5.7 ± 0.029 | 2.654 ± 0.016 |
6.259 ± 0.026 | 8.543 ± 0.029 |
5.407 ± 0.025 | 7.915 ± 0.028 |
1.122 ± 0.009 | 2.134 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
