
Hypocrea atroviridis (strain ATCC 20476 / IMI 206040) (Trichoderma atroviride)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Hypocreaceae; Trichoderma; Trichoderma atroviride
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
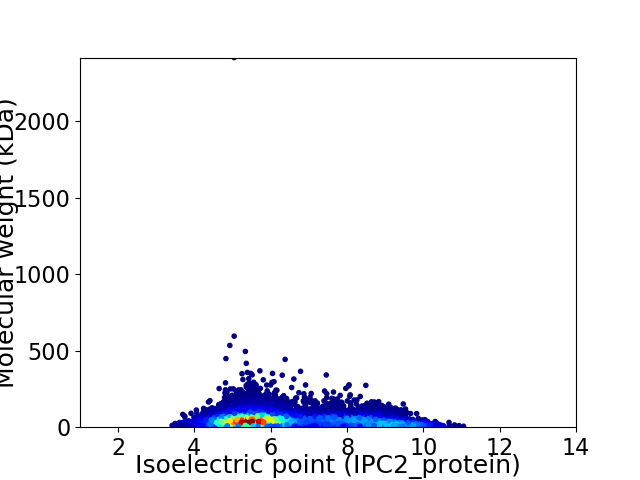
Virtual 2D-PAGE plot for 11815 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G9NZ68|G9NZ68_HYPAI MFS domain-containing protein OS=Hypocrea atroviridis (strain ATCC 20476 / IMI 206040) OX=452589 GN=TRIATDRAFT_79036 PE=3 SV=1
MM1 pKa = 7.47AVLSKK6 pKa = 10.39LLPVAAVMFGCVNPATALAVPEE28 pKa = 4.43RR29 pKa = 11.84GQRR32 pKa = 11.84WDD34 pKa = 3.17VHH36 pKa = 5.04YY37 pKa = 10.55HH38 pKa = 4.75VSLNSRR44 pKa = 11.84LNSKK48 pKa = 9.33GFPPMPHH55 pKa = 5.76EE56 pKa = 4.68AGRR59 pKa = 11.84NASAVLAEE67 pKa = 4.56RR68 pKa = 11.84GEE70 pKa = 4.19EE71 pKa = 3.88HH72 pKa = 7.66DD73 pKa = 5.35ADD75 pKa = 4.87CDD77 pKa = 4.27CQVHH81 pKa = 5.39TVILTQKK88 pKa = 10.44DD89 pKa = 3.37YY90 pKa = 11.72DD91 pKa = 4.36RR92 pKa = 11.84IQSEE96 pKa = 5.13GEE98 pKa = 4.08GHH100 pKa = 7.23PSPSTHH106 pKa = 6.14GKK108 pKa = 7.52TSEE111 pKa = 4.05NGNHH115 pKa = 5.92NEE117 pKa = 3.98RR118 pKa = 11.84GGSGEE123 pKa = 4.03YY124 pKa = 10.16KK125 pKa = 10.49EE126 pKa = 4.23SAKK129 pKa = 10.78YY130 pKa = 10.36YY131 pKa = 10.4SEE133 pKa = 4.44NGTSGEE139 pKa = 4.0YY140 pKa = 10.08RR141 pKa = 11.84EE142 pKa = 4.21SAKK145 pKa = 10.96YY146 pKa = 10.05SDD148 pKa = 3.8NGASVEE154 pKa = 4.0HH155 pKa = 6.67RR156 pKa = 11.84ASGKK160 pKa = 10.08YY161 pKa = 9.81GGSGNHH167 pKa = 5.76YY168 pKa = 10.87EE169 pKa = 3.97NGTYY173 pKa = 10.3GGSGNHH179 pKa = 5.75YY180 pKa = 10.87EE181 pKa = 3.97NGTYY185 pKa = 10.26GGSGRR190 pKa = 11.84YY191 pKa = 8.53EE192 pKa = 3.85EE193 pKa = 4.2NDD195 pKa = 3.1NYY197 pKa = 11.18GEE199 pKa = 4.07SGRR202 pKa = 11.84YY203 pKa = 8.38EE204 pKa = 4.13EE205 pKa = 4.37NDD207 pKa = 3.1NYY209 pKa = 11.18GEE211 pKa = 4.02SGRR214 pKa = 11.84YY215 pKa = 8.43EE216 pKa = 4.19EE217 pKa = 4.99IGQHH221 pKa = 6.27GEE223 pKa = 3.27IDD225 pKa = 3.81NYY227 pKa = 9.53GGRR230 pKa = 11.84YY231 pKa = 9.1KK232 pKa = 10.7EE233 pKa = 3.95VGKK236 pKa = 11.06YY237 pKa = 8.86GANGNHH243 pKa = 6.66GEE245 pKa = 4.05NEE247 pKa = 4.22KK248 pKa = 11.13YY249 pKa = 10.74GEE251 pKa = 4.1NGNYY255 pKa = 9.71GDD257 pKa = 3.51NGYY260 pKa = 10.29YY261 pKa = 10.18GKK263 pKa = 9.99NVDD266 pKa = 4.81GIADD270 pKa = 3.85QGPPGPPGPQGPPGPPGPQGNRR292 pKa = 11.84GPQGYY297 pKa = 7.63TGTKK301 pKa = 9.72GDD303 pKa = 4.05QGTQGPQGLTGSQGPAGSRR322 pKa = 11.84GPQGFTGPKK331 pKa = 9.26GDD333 pKa = 3.88QGIQGFPGATGSQGPQGNRR352 pKa = 11.84GPQGFTGPKK361 pKa = 9.63GDD363 pKa = 4.45QGNQGPQGLTGSQGPAGNRR382 pKa = 11.84GPQGLTGPAGPTGADD397 pKa = 3.34GAVGPVGPVGPVGPVGPAGADD418 pKa = 3.45GADD421 pKa = 3.82GAVGPAGPAGADD433 pKa = 3.55GAVGPVGPVGPVGPAGADD451 pKa = 3.45GADD454 pKa = 3.82GAVGPAGPAGADD466 pKa = 3.55GAVGPVGPAGADD478 pKa = 3.49GAVGPTGDD486 pKa = 4.83AGPEE490 pKa = 4.33GPIGPAGPAGADD502 pKa = 3.66GAVGPAGPVGPTGDD516 pKa = 4.35AGPEE520 pKa = 4.26GPAGADD526 pKa = 3.45GAVGPVGPAGPAGDD540 pKa = 3.95VGPTGDD546 pKa = 4.82AGPEE550 pKa = 4.43GPAGPAGPAGADD562 pKa = 3.54GAVGPTGADD571 pKa = 3.36GADD574 pKa = 3.9GADD577 pKa = 4.07GAPGPAGPAGPAGPAGADD595 pKa = 3.38GAVGPEE601 pKa = 4.47GPIGPAGADD610 pKa = 3.56GTDD613 pKa = 3.65GADD616 pKa = 3.59GAVGPAGPAGPIGPAGPAGADD637 pKa = 3.66GAVGPAGPAGADD649 pKa = 3.55GAVGPVGPAGPVGPTGDD666 pKa = 4.35AGPEE670 pKa = 4.26GPAGADD676 pKa = 3.54GAVGPAGPAGPAGDD690 pKa = 4.18VGPTGDD696 pKa = 4.89PGPEE700 pKa = 4.26GPAGPAGADD709 pKa = 3.66GAVGPAGPVGPAGDD723 pKa = 3.77VGPAGDD729 pKa = 5.0AGPEE733 pKa = 4.45GPAGPAGADD742 pKa = 3.66GAVGPAGPAGADD754 pKa = 3.51GADD757 pKa = 3.76GADD760 pKa = 3.77GAVGPAGPAGPAGPAGPAGADD781 pKa = 3.63GTDD784 pKa = 3.63GTDD787 pKa = 3.49GAVGPAGPAGPAGADD802 pKa = 3.66GAVGPAGPAGPAGADD817 pKa = 3.58GTDD820 pKa = 3.65GADD823 pKa = 3.59GAVGPAGPAGPAGADD838 pKa = 3.66GAVGPAGPAGADD850 pKa = 3.66GAVGPAGPAGPTGPTGPQGPQGPQGDD876 pKa = 3.94IADD879 pKa = 4.19YY880 pKa = 10.91DD881 pKa = 4.03YY882 pKa = 11.44LSCFTSPGDD891 pKa = 3.79STNGLAAPFATFVEE905 pKa = 5.05TVSIDD910 pKa = 3.64DD911 pKa = 4.96CATQCATIRR920 pKa = 11.84PGNPALYY927 pKa = 10.14FSLATVAGDD936 pKa = 4.04SVCSCGDD943 pKa = 3.4ALAADD948 pKa = 4.07ATANDD953 pKa = 3.73GHH955 pKa = 6.61NICNTQCTFTGQSGRR970 pKa = 11.84PVFCGGAGVVSVFASVV986 pKa = 2.7
MM1 pKa = 7.47AVLSKK6 pKa = 10.39LLPVAAVMFGCVNPATALAVPEE28 pKa = 4.43RR29 pKa = 11.84GQRR32 pKa = 11.84WDD34 pKa = 3.17VHH36 pKa = 5.04YY37 pKa = 10.55HH38 pKa = 4.75VSLNSRR44 pKa = 11.84LNSKK48 pKa = 9.33GFPPMPHH55 pKa = 5.76EE56 pKa = 4.68AGRR59 pKa = 11.84NASAVLAEE67 pKa = 4.56RR68 pKa = 11.84GEE70 pKa = 4.19EE71 pKa = 3.88HH72 pKa = 7.66DD73 pKa = 5.35ADD75 pKa = 4.87CDD77 pKa = 4.27CQVHH81 pKa = 5.39TVILTQKK88 pKa = 10.44DD89 pKa = 3.37YY90 pKa = 11.72DD91 pKa = 4.36RR92 pKa = 11.84IQSEE96 pKa = 5.13GEE98 pKa = 4.08GHH100 pKa = 7.23PSPSTHH106 pKa = 6.14GKK108 pKa = 7.52TSEE111 pKa = 4.05NGNHH115 pKa = 5.92NEE117 pKa = 3.98RR118 pKa = 11.84GGSGEE123 pKa = 4.03YY124 pKa = 10.16KK125 pKa = 10.49EE126 pKa = 4.23SAKK129 pKa = 10.78YY130 pKa = 10.36YY131 pKa = 10.4SEE133 pKa = 4.44NGTSGEE139 pKa = 4.0YY140 pKa = 10.08RR141 pKa = 11.84EE142 pKa = 4.21SAKK145 pKa = 10.96YY146 pKa = 10.05SDD148 pKa = 3.8NGASVEE154 pKa = 4.0HH155 pKa = 6.67RR156 pKa = 11.84ASGKK160 pKa = 10.08YY161 pKa = 9.81GGSGNHH167 pKa = 5.76YY168 pKa = 10.87EE169 pKa = 3.97NGTYY173 pKa = 10.3GGSGNHH179 pKa = 5.75YY180 pKa = 10.87EE181 pKa = 3.97NGTYY185 pKa = 10.26GGSGRR190 pKa = 11.84YY191 pKa = 8.53EE192 pKa = 3.85EE193 pKa = 4.2NDD195 pKa = 3.1NYY197 pKa = 11.18GEE199 pKa = 4.07SGRR202 pKa = 11.84YY203 pKa = 8.38EE204 pKa = 4.13EE205 pKa = 4.37NDD207 pKa = 3.1NYY209 pKa = 11.18GEE211 pKa = 4.02SGRR214 pKa = 11.84YY215 pKa = 8.43EE216 pKa = 4.19EE217 pKa = 4.99IGQHH221 pKa = 6.27GEE223 pKa = 3.27IDD225 pKa = 3.81NYY227 pKa = 9.53GGRR230 pKa = 11.84YY231 pKa = 9.1KK232 pKa = 10.7EE233 pKa = 3.95VGKK236 pKa = 11.06YY237 pKa = 8.86GANGNHH243 pKa = 6.66GEE245 pKa = 4.05NEE247 pKa = 4.22KK248 pKa = 11.13YY249 pKa = 10.74GEE251 pKa = 4.1NGNYY255 pKa = 9.71GDD257 pKa = 3.51NGYY260 pKa = 10.29YY261 pKa = 10.18GKK263 pKa = 9.99NVDD266 pKa = 4.81GIADD270 pKa = 3.85QGPPGPPGPQGPPGPPGPQGNRR292 pKa = 11.84GPQGYY297 pKa = 7.63TGTKK301 pKa = 9.72GDD303 pKa = 4.05QGTQGPQGLTGSQGPAGSRR322 pKa = 11.84GPQGFTGPKK331 pKa = 9.26GDD333 pKa = 3.88QGIQGFPGATGSQGPQGNRR352 pKa = 11.84GPQGFTGPKK361 pKa = 9.63GDD363 pKa = 4.45QGNQGPQGLTGSQGPAGNRR382 pKa = 11.84GPQGLTGPAGPTGADD397 pKa = 3.34GAVGPVGPVGPVGPVGPAGADD418 pKa = 3.45GADD421 pKa = 3.82GAVGPAGPAGADD433 pKa = 3.55GAVGPVGPVGPVGPAGADD451 pKa = 3.45GADD454 pKa = 3.82GAVGPAGPAGADD466 pKa = 3.55GAVGPVGPAGADD478 pKa = 3.49GAVGPTGDD486 pKa = 4.83AGPEE490 pKa = 4.33GPIGPAGPAGADD502 pKa = 3.66GAVGPAGPVGPTGDD516 pKa = 4.35AGPEE520 pKa = 4.26GPAGADD526 pKa = 3.45GAVGPVGPAGPAGDD540 pKa = 3.95VGPTGDD546 pKa = 4.82AGPEE550 pKa = 4.43GPAGPAGPAGADD562 pKa = 3.54GAVGPTGADD571 pKa = 3.36GADD574 pKa = 3.9GADD577 pKa = 4.07GAPGPAGPAGPAGPAGADD595 pKa = 3.38GAVGPEE601 pKa = 4.47GPIGPAGADD610 pKa = 3.56GTDD613 pKa = 3.65GADD616 pKa = 3.59GAVGPAGPAGPIGPAGPAGADD637 pKa = 3.66GAVGPAGPAGADD649 pKa = 3.55GAVGPVGPAGPVGPTGDD666 pKa = 4.35AGPEE670 pKa = 4.26GPAGADD676 pKa = 3.54GAVGPAGPAGPAGDD690 pKa = 4.18VGPTGDD696 pKa = 4.89PGPEE700 pKa = 4.26GPAGPAGADD709 pKa = 3.66GAVGPAGPVGPAGDD723 pKa = 3.77VGPAGDD729 pKa = 5.0AGPEE733 pKa = 4.45GPAGPAGADD742 pKa = 3.66GAVGPAGPAGADD754 pKa = 3.51GADD757 pKa = 3.76GADD760 pKa = 3.77GAVGPAGPAGPAGPAGPAGADD781 pKa = 3.63GTDD784 pKa = 3.63GTDD787 pKa = 3.49GAVGPAGPAGPAGADD802 pKa = 3.66GAVGPAGPAGPAGADD817 pKa = 3.58GTDD820 pKa = 3.65GADD823 pKa = 3.59GAVGPAGPAGPAGADD838 pKa = 3.66GAVGPAGPAGADD850 pKa = 3.66GAVGPAGPAGPTGPTGPQGPQGPQGDD876 pKa = 3.94IADD879 pKa = 4.19YY880 pKa = 10.91DD881 pKa = 4.03YY882 pKa = 11.44LSCFTSPGDD891 pKa = 3.79STNGLAAPFATFVEE905 pKa = 5.05TVSIDD910 pKa = 3.64DD911 pKa = 4.96CATQCATIRR920 pKa = 11.84PGNPALYY927 pKa = 10.14FSLATVAGDD936 pKa = 4.04SVCSCGDD943 pKa = 3.4ALAADD948 pKa = 4.07ATANDD953 pKa = 3.73GHH955 pKa = 6.61NICNTQCTFTGQSGRR970 pKa = 11.84PVFCGGAGVVSVFASVV986 pKa = 2.7
Molecular weight: 90.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G9NJ98|G9NJ98_HYPAI Protein kish OS=Hypocrea atroviridis (strain ATCC 20476 / IMI 206040) OX=452589 GN=TRIATDRAFT_255048 PE=3 SV=1
MM1 pKa = 7.87PSHH4 pKa = 5.85KK5 pKa = 9.4TFRR8 pKa = 11.84TKK10 pKa = 10.46QKK12 pKa = 9.86LAKK15 pKa = 9.55AQKK18 pKa = 8.93QNRR21 pKa = 11.84PVPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.14WRR45 pKa = 11.84KK46 pKa = 7.41TRR48 pKa = 11.84LGII51 pKa = 4.46
MM1 pKa = 7.87PSHH4 pKa = 5.85KK5 pKa = 9.4TFRR8 pKa = 11.84TKK10 pKa = 10.46QKK12 pKa = 9.86LAKK15 pKa = 9.55AQKK18 pKa = 8.93QNRR21 pKa = 11.84PVPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.14WRR45 pKa = 11.84KK46 pKa = 7.41TRR48 pKa = 11.84LGII51 pKa = 4.46
Molecular weight: 6.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5388752 |
43 |
21901 |
456.1 |
50.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.836 ± 0.019 | 1.277 ± 0.009 |
5.763 ± 0.016 | 6.083 ± 0.02 |
3.815 ± 0.015 | 6.804 ± 0.025 |
2.365 ± 0.009 | 5.171 ± 0.015 |
4.932 ± 0.022 | 9.028 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.218 ± 0.009 | 3.76 ± 0.011 |
5.724 ± 0.024 | 4.055 ± 0.018 |
5.876 ± 0.018 | 8.199 ± 0.024 |
5.729 ± 0.015 | 6.038 ± 0.017 |
1.509 ± 0.008 | 2.817 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
