
Cyprinid herpesvirus 1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Alloherpesviridae; Cyprinivirus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
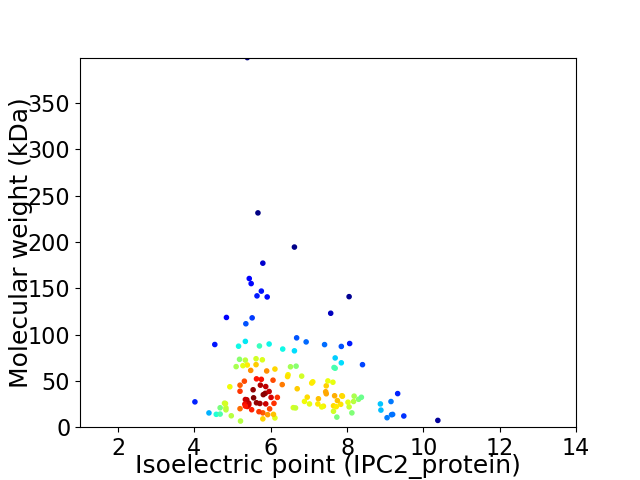
Virtual 2D-PAGE plot for 137 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
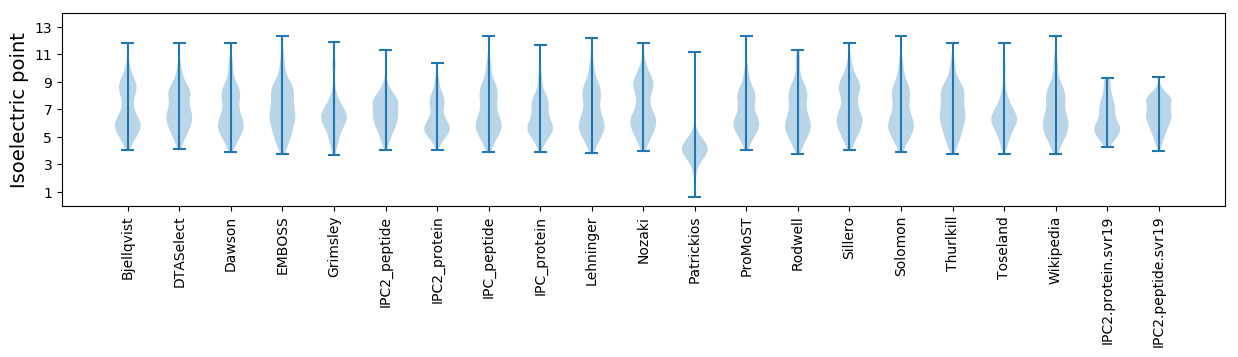
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7PC75|K7PC75_9VIRU Protein ORF70 OS=Cyprinid herpesvirus 1 OX=317858 GN=CyHV1_ORF70 PE=4 SV=1
MM1 pKa = 7.51ALLWSLVFLLATCAISVTCQSVTWTRR27 pKa = 11.84SSPCSPMGTTLVITGAYY44 pKa = 7.87TMPTGQYY51 pKa = 9.84AIAGAILSPNMDD63 pKa = 3.33IKK65 pKa = 10.16TVATVTTRR73 pKa = 11.84ATVNLTPGNKK83 pKa = 8.56GTVTFTLSGVRR94 pKa = 11.84MSDD97 pKa = 2.76ATFTGFALKK106 pKa = 10.81VLTTTGASTTMSTLAPPITVATNPSIRR133 pKa = 11.84VLRR136 pKa = 11.84YY137 pKa = 9.26NDD139 pKa = 3.32YY140 pKa = 10.82SLGGGTVLRR149 pKa = 11.84CVQSCNSTINNHH161 pKa = 3.84VWYY164 pKa = 10.64KK165 pKa = 10.62NGLKK169 pKa = 9.9IGSAIGTDD177 pKa = 3.38MTAVDD182 pKa = 3.98SGLYY186 pKa = 8.53TCEE189 pKa = 4.06PDD191 pKa = 3.75GSINTRR197 pKa = 11.84SNSIWVGTPTYY208 pKa = 10.79DD209 pKa = 5.0CPFDD213 pKa = 3.71LAKK216 pKa = 9.68WCSSGFITRR225 pKa = 11.84RR226 pKa = 11.84HH227 pKa = 5.28EE228 pKa = 4.59NNVKK232 pKa = 10.32AYY234 pKa = 10.43NSDD237 pKa = 3.16HH238 pKa = 6.09TRR240 pKa = 11.84TVGNTFVCGSSTDD253 pKa = 3.56GTSPAISTRR262 pKa = 11.84GMCPNRR268 pKa = 11.84TTCTSEE274 pKa = 3.79KK275 pKa = 8.61TTMCANNNPVINVACGMVISYY296 pKa = 7.56TCPTISPFEE305 pKa = 4.39SIGIVSSSGSEE316 pKa = 3.94FALSPVAGEE325 pKa = 4.26RR326 pKa = 11.84RR327 pKa = 11.84LEE329 pKa = 4.04TSTKK333 pKa = 9.87GNVYY337 pKa = 10.23KK338 pKa = 10.39IWCGSDD344 pKa = 3.41YY345 pKa = 11.25EE346 pKa = 4.81LVAPCGTSSSYY357 pKa = 10.65SAHH360 pKa = 7.14PSTFIPIGAGATWTTTGVTQSEE382 pKa = 4.49TMIIAVGTVFNITCPDD398 pKa = 3.34NTVVAWYY405 pKa = 10.08KK406 pKa = 11.11DD407 pKa = 3.18KK408 pKa = 10.64TLFSSLIGSRR418 pKa = 11.84STKK421 pKa = 10.94NMVKK425 pKa = 10.43NLTADD430 pKa = 3.6DD431 pKa = 4.01YY432 pKa = 12.18GMWEE436 pKa = 4.48CVSFTTTGSTLGARR450 pKa = 11.84TRR452 pKa = 11.84LISISAPTTTVTTPLTTPSTTAATTAAATNATTAATTAAATNATTAATTAAATNATTAATTAATTAAATNATTAATTEE530 pKa = 4.0AATNATTAATTAATTEE546 pKa = 4.0AATNATTAATTASATNATTAATTEE570 pKa = 4.0AATNATTAATTEE582 pKa = 4.0AATNATTAATTEE594 pKa = 4.0AATNATTAATTAATTEE610 pKa = 4.0AATNATTAATTAATTEE626 pKa = 4.0AATNATTAATTAATTEE642 pKa = 4.0AATNATTAATTAATTEE658 pKa = 4.0AATNATTAATTAATTEE674 pKa = 4.0AATNATTAATTAATTEE690 pKa = 4.0AATNATTEE698 pKa = 4.01AATNATTEE706 pKa = 4.01AATNATMSPTTPDD719 pKa = 3.06ASNATTDD726 pKa = 3.32ASTPDD731 pKa = 3.28ASNATTAATTEE742 pKa = 4.26DD743 pKa = 3.42ATNATTEE750 pKa = 3.97AATNATMSPATPDD763 pKa = 3.2ASNATTDD770 pKa = 3.32ASTPDD775 pKa = 3.28ASNATTAATTPDD787 pKa = 3.54ASNATTDD794 pKa = 3.3AATNATMSPTTPDD807 pKa = 3.06ASNATTDD814 pKa = 3.12ASTPDD819 pKa = 3.48TSNATTDD826 pKa = 2.83ATTAGNTPATTPGTPEE842 pKa = 3.96TTTGRR847 pKa = 11.84PLTTPSPTTQSTPINPTGLSTLTMAAIIGGAAGGAALIAAIVAVCVIHH895 pKa = 7.04NGII898 pKa = 4.04
MM1 pKa = 7.51ALLWSLVFLLATCAISVTCQSVTWTRR27 pKa = 11.84SSPCSPMGTTLVITGAYY44 pKa = 7.87TMPTGQYY51 pKa = 9.84AIAGAILSPNMDD63 pKa = 3.33IKK65 pKa = 10.16TVATVTTRR73 pKa = 11.84ATVNLTPGNKK83 pKa = 8.56GTVTFTLSGVRR94 pKa = 11.84MSDD97 pKa = 2.76ATFTGFALKK106 pKa = 10.81VLTTTGASTTMSTLAPPITVATNPSIRR133 pKa = 11.84VLRR136 pKa = 11.84YY137 pKa = 9.26NDD139 pKa = 3.32YY140 pKa = 10.82SLGGGTVLRR149 pKa = 11.84CVQSCNSTINNHH161 pKa = 3.84VWYY164 pKa = 10.64KK165 pKa = 10.62NGLKK169 pKa = 9.9IGSAIGTDD177 pKa = 3.38MTAVDD182 pKa = 3.98SGLYY186 pKa = 8.53TCEE189 pKa = 4.06PDD191 pKa = 3.75GSINTRR197 pKa = 11.84SNSIWVGTPTYY208 pKa = 10.79DD209 pKa = 5.0CPFDD213 pKa = 3.71LAKK216 pKa = 9.68WCSSGFITRR225 pKa = 11.84RR226 pKa = 11.84HH227 pKa = 5.28EE228 pKa = 4.59NNVKK232 pKa = 10.32AYY234 pKa = 10.43NSDD237 pKa = 3.16HH238 pKa = 6.09TRR240 pKa = 11.84TVGNTFVCGSSTDD253 pKa = 3.56GTSPAISTRR262 pKa = 11.84GMCPNRR268 pKa = 11.84TTCTSEE274 pKa = 3.79KK275 pKa = 8.61TTMCANNNPVINVACGMVISYY296 pKa = 7.56TCPTISPFEE305 pKa = 4.39SIGIVSSSGSEE316 pKa = 3.94FALSPVAGEE325 pKa = 4.26RR326 pKa = 11.84RR327 pKa = 11.84LEE329 pKa = 4.04TSTKK333 pKa = 9.87GNVYY337 pKa = 10.23KK338 pKa = 10.39IWCGSDD344 pKa = 3.41YY345 pKa = 11.25EE346 pKa = 4.81LVAPCGTSSSYY357 pKa = 10.65SAHH360 pKa = 7.14PSTFIPIGAGATWTTTGVTQSEE382 pKa = 4.49TMIIAVGTVFNITCPDD398 pKa = 3.34NTVVAWYY405 pKa = 10.08KK406 pKa = 11.11DD407 pKa = 3.18KK408 pKa = 10.64TLFSSLIGSRR418 pKa = 11.84STKK421 pKa = 10.94NMVKK425 pKa = 10.43NLTADD430 pKa = 3.6DD431 pKa = 4.01YY432 pKa = 12.18GMWEE436 pKa = 4.48CVSFTTTGSTLGARR450 pKa = 11.84TRR452 pKa = 11.84LISISAPTTTVTTPLTTPSTTAATTAAATNATTAATTAAATNATTAATTAAATNATTAATTAATTAAATNATTAATTEE530 pKa = 4.0AATNATTAATTAATTEE546 pKa = 4.0AATNATTAATTASATNATTAATTEE570 pKa = 4.0AATNATTAATTEE582 pKa = 4.0AATNATTAATTEE594 pKa = 4.0AATNATTAATTAATTEE610 pKa = 4.0AATNATTAATTAATTEE626 pKa = 4.0AATNATTAATTAATTEE642 pKa = 4.0AATNATTAATTAATTEE658 pKa = 4.0AATNATTAATTAATTEE674 pKa = 4.0AATNATTAATTAATTEE690 pKa = 4.0AATNATTEE698 pKa = 4.01AATNATTEE706 pKa = 4.01AATNATMSPTTPDD719 pKa = 3.06ASNATTDD726 pKa = 3.32ASTPDD731 pKa = 3.28ASNATTAATTEE742 pKa = 4.26DD743 pKa = 3.42ATNATTEE750 pKa = 3.97AATNATMSPATPDD763 pKa = 3.2ASNATTDD770 pKa = 3.32ASTPDD775 pKa = 3.28ASNATTAATTPDD787 pKa = 3.54ASNATTDD794 pKa = 3.3AATNATMSPTTPDD807 pKa = 3.06ASNATTDD814 pKa = 3.12ASTPDD819 pKa = 3.48TSNATTDD826 pKa = 2.83ATTAGNTPATTPGTPEE842 pKa = 3.96TTTGRR847 pKa = 11.84PLTTPSPTTQSTPINPTGLSTLTMAAIIGGAAGGAALIAAIVAVCVIHH895 pKa = 7.04NGII898 pKa = 4.04
Molecular weight: 89.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7PC85|K7PC85_9VIRU Protein ORF85 OS=Cyprinid herpesvirus 1 OX=317858 GN=CyHV1_ORF85 PE=4 SV=1
MM1 pKa = 7.6EE2 pKa = 5.8LPRR5 pKa = 11.84VAIPAEE11 pKa = 3.94ARR13 pKa = 11.84EE14 pKa = 4.04LGITKK19 pKa = 8.84TLSEE23 pKa = 5.39LYY25 pKa = 9.43PSHH28 pKa = 7.01RR29 pKa = 11.84FYY31 pKa = 11.06RR32 pKa = 11.84GQRR35 pKa = 11.84RR36 pKa = 11.84IAIKK40 pKa = 10.82LEE42 pKa = 4.02VEE44 pKa = 4.05GDD46 pKa = 3.69EE47 pKa = 4.01QDD49 pKa = 3.78LVVRR53 pKa = 11.84LTKK56 pKa = 10.09SLRR59 pKa = 11.84GRR61 pKa = 11.84VWTKK65 pKa = 9.26PQLRR69 pKa = 11.84AQILSLLPGLSGQQGEE85 pKa = 4.78DD86 pKa = 3.0ACQAAAEE93 pKa = 4.24ALTRR97 pKa = 11.84SAPFTLKK104 pKa = 10.2SVRR107 pKa = 11.84SALISTRR114 pKa = 11.84FFARR118 pKa = 11.84MGSLVDD124 pKa = 3.54SRR126 pKa = 11.84VKK128 pKa = 10.77RR129 pKa = 11.84EE130 pKa = 3.75FEE132 pKa = 3.97CGKK135 pKa = 10.79GYY137 pKa = 10.65LVDD140 pKa = 3.81HH141 pKa = 6.75LKK143 pKa = 10.66AVYY146 pKa = 9.29GWRR149 pKa = 11.84PLASAVNVTSKK160 pKa = 10.86VPLGHH165 pKa = 5.75SCEE168 pKa = 4.35RR169 pKa = 11.84CHH171 pKa = 6.98SVCSRR176 pKa = 11.84QVPSHH181 pKa = 5.44KK182 pKa = 9.65TEE184 pKa = 4.17LDD186 pKa = 3.11AVAVDD191 pKa = 3.74EE192 pKa = 4.59RR193 pKa = 11.84GEE195 pKa = 4.1YY196 pKa = 10.82ALLEE200 pKa = 4.22IKK202 pKa = 9.31TRR204 pKa = 11.84SSSTVTAALLRR215 pKa = 11.84RR216 pKa = 11.84YY217 pKa = 8.5QVQTWIGEE225 pKa = 4.02MMFRR229 pKa = 11.84NTYY232 pKa = 9.92GMCTSKK238 pKa = 10.76AIHH241 pKa = 6.37SYY243 pKa = 10.37IVFVDD248 pKa = 4.0PCSYY252 pKa = 10.36TVDD255 pKa = 3.35RR256 pKa = 11.84VICVDD261 pKa = 3.53SKK263 pKa = 11.63LNRR266 pKa = 11.84PSPRR270 pKa = 11.84LFALFPSLLNLCQAKK285 pKa = 10.38LSAPRR290 pKa = 11.84VLLRR294 pKa = 11.84PPAGRR299 pKa = 11.84PSRR302 pKa = 11.84PCASGAAVKK311 pKa = 10.41RR312 pKa = 11.84KK313 pKa = 9.69RR314 pKa = 11.84PSGAAAAEE322 pKa = 4.54KK323 pKa = 10.19KK324 pKa = 10.33RR325 pKa = 11.84KK326 pKa = 9.07AA327 pKa = 3.43
MM1 pKa = 7.6EE2 pKa = 5.8LPRR5 pKa = 11.84VAIPAEE11 pKa = 3.94ARR13 pKa = 11.84EE14 pKa = 4.04LGITKK19 pKa = 8.84TLSEE23 pKa = 5.39LYY25 pKa = 9.43PSHH28 pKa = 7.01RR29 pKa = 11.84FYY31 pKa = 11.06RR32 pKa = 11.84GQRR35 pKa = 11.84RR36 pKa = 11.84IAIKK40 pKa = 10.82LEE42 pKa = 4.02VEE44 pKa = 4.05GDD46 pKa = 3.69EE47 pKa = 4.01QDD49 pKa = 3.78LVVRR53 pKa = 11.84LTKK56 pKa = 10.09SLRR59 pKa = 11.84GRR61 pKa = 11.84VWTKK65 pKa = 9.26PQLRR69 pKa = 11.84AQILSLLPGLSGQQGEE85 pKa = 4.78DD86 pKa = 3.0ACQAAAEE93 pKa = 4.24ALTRR97 pKa = 11.84SAPFTLKK104 pKa = 10.2SVRR107 pKa = 11.84SALISTRR114 pKa = 11.84FFARR118 pKa = 11.84MGSLVDD124 pKa = 3.54SRR126 pKa = 11.84VKK128 pKa = 10.77RR129 pKa = 11.84EE130 pKa = 3.75FEE132 pKa = 3.97CGKK135 pKa = 10.79GYY137 pKa = 10.65LVDD140 pKa = 3.81HH141 pKa = 6.75LKK143 pKa = 10.66AVYY146 pKa = 9.29GWRR149 pKa = 11.84PLASAVNVTSKK160 pKa = 10.86VPLGHH165 pKa = 5.75SCEE168 pKa = 4.35RR169 pKa = 11.84CHH171 pKa = 6.98SVCSRR176 pKa = 11.84QVPSHH181 pKa = 5.44KK182 pKa = 9.65TEE184 pKa = 4.17LDD186 pKa = 3.11AVAVDD191 pKa = 3.74EE192 pKa = 4.59RR193 pKa = 11.84GEE195 pKa = 4.1YY196 pKa = 10.82ALLEE200 pKa = 4.22IKK202 pKa = 9.31TRR204 pKa = 11.84SSSTVTAALLRR215 pKa = 11.84RR216 pKa = 11.84YY217 pKa = 8.5QVQTWIGEE225 pKa = 4.02MMFRR229 pKa = 11.84NTYY232 pKa = 9.92GMCTSKK238 pKa = 10.76AIHH241 pKa = 6.37SYY243 pKa = 10.37IVFVDD248 pKa = 4.0PCSYY252 pKa = 10.36TVDD255 pKa = 3.35RR256 pKa = 11.84VICVDD261 pKa = 3.53SKK263 pKa = 11.63LNRR266 pKa = 11.84PSPRR270 pKa = 11.84LFALFPSLLNLCQAKK285 pKa = 10.38LSAPRR290 pKa = 11.84VLLRR294 pKa = 11.84PPAGRR299 pKa = 11.84PSRR302 pKa = 11.84PCASGAAVKK311 pKa = 10.41RR312 pKa = 11.84KK313 pKa = 9.69RR314 pKa = 11.84PSGAAAAEE322 pKa = 4.54KK323 pKa = 10.19KK324 pKa = 10.33RR325 pKa = 11.84KK326 pKa = 9.07AA327 pKa = 3.43
Molecular weight: 36.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
65611 |
64 |
3611 |
478.9 |
53.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.241 ± 0.257 | 2.55 ± 0.116 |
5.394 ± 0.11 | 6.243 ± 0.211 |
3.51 ± 0.126 | 5.587 ± 0.118 |
2.401 ± 0.092 | 3.824 ± 0.108 |
4.928 ± 0.173 | 9.128 ± 0.186 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.716 ± 0.087 | 3.64 ± 0.153 |
5.883 ± 0.212 | 3.597 ± 0.113 |
5.991 ± 0.195 | 8.092 ± 0.172 |
6.776 ± 0.323 | 6.909 ± 0.132 |
1.468 ± 0.075 | 3.123 ± 0.094 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
