
Avian infectious bursal disease virus (strain Chicken/Cuba/Soroa/1998) (IBDV) (Gumboro disease virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Birnaviridae; Avibirnavirus; Infectious bursal disease virus
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
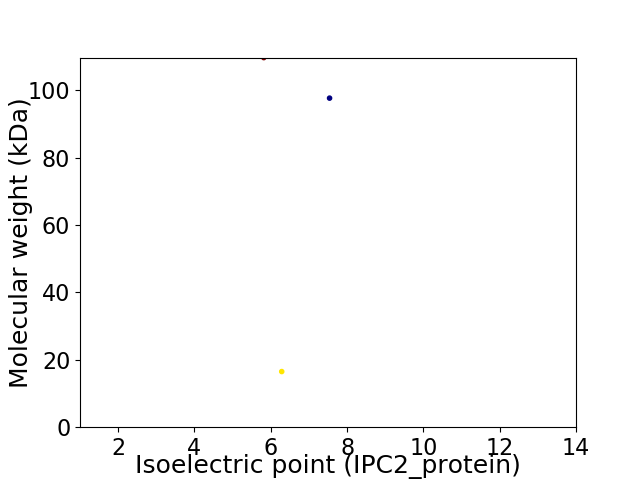
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9WI42|POLS_IBDVB Structural polyprotein OS=Avian infectious bursal disease virus (strain Chicken/Cuba/Soroa/1998) OX=645118 PE=1 SV=1
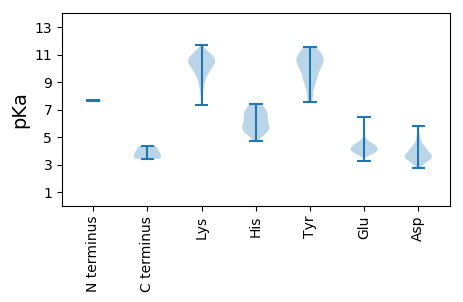
MM1 pKa = 7.63TNLSDD6 pKa = 3.41QTQQIVPFIRR16 pKa = 11.84SLLMPTTGPASIPDD30 pKa = 3.64DD31 pKa = 3.68TLEE34 pKa = 3.92KK35 pKa = 9.68HH36 pKa = 5.47TLRR39 pKa = 11.84SEE41 pKa = 3.92TSTYY45 pKa = 10.97NLTVGDD51 pKa = 4.14TGSGLIVFFPGFPGSIVGAHH71 pKa = 5.48YY72 pKa = 9.25TLQGNGNYY80 pKa = 10.14KK81 pKa = 9.78FDD83 pKa = 3.92QMLLTAQNLPASYY96 pKa = 10.21NYY98 pKa = 9.44CRR100 pKa = 11.84LVSRR104 pKa = 11.84SLTVRR109 pKa = 11.84SSTLPGGVYY118 pKa = 10.67ALNGTINAVTFQGSLSEE135 pKa = 4.27LTDD138 pKa = 3.21VSYY141 pKa = 11.52NGLMSATANINDD153 pKa = 4.22KK154 pKa = 10.59IGNVLVGEE162 pKa = 4.48GVTVLSLPTSYY173 pKa = 11.5DD174 pKa = 2.89LGYY177 pKa = 10.9VRR179 pKa = 11.84LGDD182 pKa = 4.85PIPAIGLDD190 pKa = 3.48PKK192 pKa = 10.43MVATCDD198 pKa = 3.2SSDD201 pKa = 3.38RR202 pKa = 11.84PRR204 pKa = 11.84VYY206 pKa = 10.49TITAADD212 pKa = 4.51DD213 pKa = 3.84YY214 pKa = 11.05QFSSQYY220 pKa = 10.28QPGGVTITLFSANIDD235 pKa = 4.21AITSLSVGGEE245 pKa = 3.94LVFRR249 pKa = 11.84TSVHH253 pKa = 5.56GLVLGATIYY262 pKa = 11.11LIGFDD267 pKa = 3.63GTTVITRR274 pKa = 11.84AVAANNGLTTGTDD287 pKa = 2.87NLMPFNLVIPTNEE300 pKa = 3.25ITQPITSIKK309 pKa = 10.6LEE311 pKa = 4.0IVTSKK316 pKa = 10.91SGGQAGDD323 pKa = 3.41QMSWSARR330 pKa = 11.84GSLAVTIHH338 pKa = 6.06GGNYY342 pKa = 9.04PGALRR347 pKa = 11.84PVTLVAYY354 pKa = 9.1EE355 pKa = 4.18RR356 pKa = 11.84VATGSVVTVAGVSNFEE372 pKa = 5.03LIPNPEE378 pKa = 3.96LAKK381 pKa = 11.03NLVTEE386 pKa = 4.26YY387 pKa = 11.44GRR389 pKa = 11.84FDD391 pKa = 4.12PGAMNYY397 pKa = 7.66TKK399 pKa = 10.75LILSEE404 pKa = 4.11RR405 pKa = 11.84DD406 pKa = 2.95RR407 pKa = 11.84LGIKK411 pKa = 7.3TVWPTRR417 pKa = 11.84EE418 pKa = 3.79YY419 pKa = 10.7TDD421 pKa = 3.6FRR423 pKa = 11.84EE424 pKa = 4.25YY425 pKa = 10.92FMEE428 pKa = 4.29VADD431 pKa = 4.92LNSPLKK437 pKa = 10.41IAGAFGFKK445 pKa = 10.39DD446 pKa = 3.63IIRR449 pKa = 11.84AIRR452 pKa = 11.84RR453 pKa = 11.84IAVPVVSTLFPPAAPLAHH471 pKa = 7.26AIGEE475 pKa = 4.5GVDD478 pKa = 3.58YY479 pKa = 11.29LLGDD483 pKa = 3.95EE484 pKa = 4.72AQAASGTARR493 pKa = 11.84AASGKK498 pKa = 9.96ARR500 pKa = 11.84AASGRR505 pKa = 11.84IRR507 pKa = 11.84QLTLAADD514 pKa = 3.27KK515 pKa = 10.88GYY517 pKa = 10.91EE518 pKa = 4.07VVANLFQVPQNPVVDD533 pKa = 5.83GILASPGVLRR543 pKa = 11.84GAHH546 pKa = 6.39NLDD549 pKa = 3.45CVLRR553 pKa = 11.84EE554 pKa = 4.02GATLFPVVITTVEE567 pKa = 3.82DD568 pKa = 3.49AMTPKK573 pKa = 10.78ALNSKK578 pKa = 8.69MFAVIEE584 pKa = 4.28GVRR587 pKa = 11.84EE588 pKa = 3.92DD589 pKa = 4.45LQPPSQRR596 pKa = 11.84GSFIRR601 pKa = 11.84TLSGHH606 pKa = 5.86RR607 pKa = 11.84VYY609 pKa = 10.95GYY611 pKa = 11.08APDD614 pKa = 3.92GVLPLEE620 pKa = 4.29TGRR623 pKa = 11.84DD624 pKa = 3.85YY625 pKa = 11.49TVVPIDD631 pKa = 4.92DD632 pKa = 3.82VWDD635 pKa = 4.19DD636 pKa = 4.84SIMLSKK642 pKa = 11.0DD643 pKa = 3.59PIPPIVGNSGNLAIAYY659 pKa = 8.15MDD661 pKa = 3.36VFRR664 pKa = 11.84PKK666 pKa = 10.6VPIHH670 pKa = 5.55VAMTGALNACGEE682 pKa = 4.16IEE684 pKa = 4.17KK685 pKa = 10.91VSFRR689 pKa = 11.84STKK692 pKa = 10.15LATAHH697 pKa = 7.1RR698 pKa = 11.84LGLRR702 pKa = 11.84LAGPGAFDD710 pKa = 5.0VNTGPNWATFIKK722 pKa = 10.24RR723 pKa = 11.84FPHH726 pKa = 6.93NPRR729 pKa = 11.84DD730 pKa = 3.86WDD732 pKa = 3.68RR733 pKa = 11.84LPYY736 pKa = 10.78LNLPYY741 pKa = 10.53LPPNAGRR748 pKa = 11.84QYY750 pKa = 9.83HH751 pKa = 6.51LAMAASEE758 pKa = 4.14FKK760 pKa = 9.23EE761 pKa = 4.5TPEE764 pKa = 4.24LEE766 pKa = 4.34SAVRR770 pKa = 11.84AMEE773 pKa = 3.97AAANVDD779 pKa = 3.86PLFQSALSVFMWLEE793 pKa = 3.76EE794 pKa = 4.06NGIVTDD800 pKa = 3.69MANFALSDD808 pKa = 4.05PNAHH812 pKa = 7.07RR813 pKa = 11.84MRR815 pKa = 11.84NFLANAPQAGSKK827 pKa = 7.76SQRR830 pKa = 11.84AKK832 pKa = 10.94YY833 pKa = 8.02GTAGYY838 pKa = 9.76GVEE841 pKa = 4.38ARR843 pKa = 11.84GPTPEE848 pKa = 3.68EE849 pKa = 3.66AQRR852 pKa = 11.84EE853 pKa = 4.02KK854 pKa = 10.55DD855 pKa = 3.34TRR857 pKa = 11.84ISKK860 pKa = 10.43KK861 pKa = 9.06MEE863 pKa = 3.68TMGIYY868 pKa = 10.18FATPEE873 pKa = 3.79WVALNGHH880 pKa = 7.16RR881 pKa = 11.84GPSPGQVKK889 pKa = 9.23YY890 pKa = 9.19WQNKK894 pKa = 8.73RR895 pKa = 11.84EE896 pKa = 4.11IPDD899 pKa = 3.84PNEE902 pKa = 4.54DD903 pKa = 3.6YY904 pKa = 11.22LDD906 pKa = 3.79YY907 pKa = 11.55VHH909 pKa = 7.37AEE911 pKa = 3.96KK912 pKa = 10.86SRR914 pKa = 11.84LASEE918 pKa = 4.11EE919 pKa = 4.0QILRR923 pKa = 11.84AATSIYY929 pKa = 9.8GAPGQAEE936 pKa = 4.26PPQAFIDD943 pKa = 4.24EE944 pKa = 4.38VAKK947 pKa = 10.82VYY949 pKa = 10.07EE950 pKa = 4.43INHH953 pKa = 5.42GRR955 pKa = 11.84GPNQEE960 pKa = 3.86QMKK963 pKa = 10.54DD964 pKa = 3.44LLLTAMEE971 pKa = 4.32MKK973 pKa = 10.22HH974 pKa = 5.91RR975 pKa = 11.84NPRR978 pKa = 11.84RR979 pKa = 11.84ALPKK983 pKa = 10.15PKK985 pKa = 9.54PKK987 pKa = 10.28PNAPTQRR994 pKa = 11.84PPGRR998 pKa = 11.84LGRR1001 pKa = 11.84WIRR1004 pKa = 11.84TVSDD1008 pKa = 3.49EE1009 pKa = 4.72DD1010 pKa = 4.27LEE1012 pKa = 4.35
MM1 pKa = 7.63TNLSDD6 pKa = 3.41QTQQIVPFIRR16 pKa = 11.84SLLMPTTGPASIPDD30 pKa = 3.64DD31 pKa = 3.68TLEE34 pKa = 3.92KK35 pKa = 9.68HH36 pKa = 5.47TLRR39 pKa = 11.84SEE41 pKa = 3.92TSTYY45 pKa = 10.97NLTVGDD51 pKa = 4.14TGSGLIVFFPGFPGSIVGAHH71 pKa = 5.48YY72 pKa = 9.25TLQGNGNYY80 pKa = 10.14KK81 pKa = 9.78FDD83 pKa = 3.92QMLLTAQNLPASYY96 pKa = 10.21NYY98 pKa = 9.44CRR100 pKa = 11.84LVSRR104 pKa = 11.84SLTVRR109 pKa = 11.84SSTLPGGVYY118 pKa = 10.67ALNGTINAVTFQGSLSEE135 pKa = 4.27LTDD138 pKa = 3.21VSYY141 pKa = 11.52NGLMSATANINDD153 pKa = 4.22KK154 pKa = 10.59IGNVLVGEE162 pKa = 4.48GVTVLSLPTSYY173 pKa = 11.5DD174 pKa = 2.89LGYY177 pKa = 10.9VRR179 pKa = 11.84LGDD182 pKa = 4.85PIPAIGLDD190 pKa = 3.48PKK192 pKa = 10.43MVATCDD198 pKa = 3.2SSDD201 pKa = 3.38RR202 pKa = 11.84PRR204 pKa = 11.84VYY206 pKa = 10.49TITAADD212 pKa = 4.51DD213 pKa = 3.84YY214 pKa = 11.05QFSSQYY220 pKa = 10.28QPGGVTITLFSANIDD235 pKa = 4.21AITSLSVGGEE245 pKa = 3.94LVFRR249 pKa = 11.84TSVHH253 pKa = 5.56GLVLGATIYY262 pKa = 11.11LIGFDD267 pKa = 3.63GTTVITRR274 pKa = 11.84AVAANNGLTTGTDD287 pKa = 2.87NLMPFNLVIPTNEE300 pKa = 3.25ITQPITSIKK309 pKa = 10.6LEE311 pKa = 4.0IVTSKK316 pKa = 10.91SGGQAGDD323 pKa = 3.41QMSWSARR330 pKa = 11.84GSLAVTIHH338 pKa = 6.06GGNYY342 pKa = 9.04PGALRR347 pKa = 11.84PVTLVAYY354 pKa = 9.1EE355 pKa = 4.18RR356 pKa = 11.84VATGSVVTVAGVSNFEE372 pKa = 5.03LIPNPEE378 pKa = 3.96LAKK381 pKa = 11.03NLVTEE386 pKa = 4.26YY387 pKa = 11.44GRR389 pKa = 11.84FDD391 pKa = 4.12PGAMNYY397 pKa = 7.66TKK399 pKa = 10.75LILSEE404 pKa = 4.11RR405 pKa = 11.84DD406 pKa = 2.95RR407 pKa = 11.84LGIKK411 pKa = 7.3TVWPTRR417 pKa = 11.84EE418 pKa = 3.79YY419 pKa = 10.7TDD421 pKa = 3.6FRR423 pKa = 11.84EE424 pKa = 4.25YY425 pKa = 10.92FMEE428 pKa = 4.29VADD431 pKa = 4.92LNSPLKK437 pKa = 10.41IAGAFGFKK445 pKa = 10.39DD446 pKa = 3.63IIRR449 pKa = 11.84AIRR452 pKa = 11.84RR453 pKa = 11.84IAVPVVSTLFPPAAPLAHH471 pKa = 7.26AIGEE475 pKa = 4.5GVDD478 pKa = 3.58YY479 pKa = 11.29LLGDD483 pKa = 3.95EE484 pKa = 4.72AQAASGTARR493 pKa = 11.84AASGKK498 pKa = 9.96ARR500 pKa = 11.84AASGRR505 pKa = 11.84IRR507 pKa = 11.84QLTLAADD514 pKa = 3.27KK515 pKa = 10.88GYY517 pKa = 10.91EE518 pKa = 4.07VVANLFQVPQNPVVDD533 pKa = 5.83GILASPGVLRR543 pKa = 11.84GAHH546 pKa = 6.39NLDD549 pKa = 3.45CVLRR553 pKa = 11.84EE554 pKa = 4.02GATLFPVVITTVEE567 pKa = 3.82DD568 pKa = 3.49AMTPKK573 pKa = 10.78ALNSKK578 pKa = 8.69MFAVIEE584 pKa = 4.28GVRR587 pKa = 11.84EE588 pKa = 3.92DD589 pKa = 4.45LQPPSQRR596 pKa = 11.84GSFIRR601 pKa = 11.84TLSGHH606 pKa = 5.86RR607 pKa = 11.84VYY609 pKa = 10.95GYY611 pKa = 11.08APDD614 pKa = 3.92GVLPLEE620 pKa = 4.29TGRR623 pKa = 11.84DD624 pKa = 3.85YY625 pKa = 11.49TVVPIDD631 pKa = 4.92DD632 pKa = 3.82VWDD635 pKa = 4.19DD636 pKa = 4.84SIMLSKK642 pKa = 11.0DD643 pKa = 3.59PIPPIVGNSGNLAIAYY659 pKa = 8.15MDD661 pKa = 3.36VFRR664 pKa = 11.84PKK666 pKa = 10.6VPIHH670 pKa = 5.55VAMTGALNACGEE682 pKa = 4.16IEE684 pKa = 4.17KK685 pKa = 10.91VSFRR689 pKa = 11.84STKK692 pKa = 10.15LATAHH697 pKa = 7.1RR698 pKa = 11.84LGLRR702 pKa = 11.84LAGPGAFDD710 pKa = 5.0VNTGPNWATFIKK722 pKa = 10.24RR723 pKa = 11.84FPHH726 pKa = 6.93NPRR729 pKa = 11.84DD730 pKa = 3.86WDD732 pKa = 3.68RR733 pKa = 11.84LPYY736 pKa = 10.78LNLPYY741 pKa = 10.53LPPNAGRR748 pKa = 11.84QYY750 pKa = 9.83HH751 pKa = 6.51LAMAASEE758 pKa = 4.14FKK760 pKa = 9.23EE761 pKa = 4.5TPEE764 pKa = 4.24LEE766 pKa = 4.34SAVRR770 pKa = 11.84AMEE773 pKa = 3.97AAANVDD779 pKa = 3.86PLFQSALSVFMWLEE793 pKa = 3.76EE794 pKa = 4.06NGIVTDD800 pKa = 3.69MANFALSDD808 pKa = 4.05PNAHH812 pKa = 7.07RR813 pKa = 11.84MRR815 pKa = 11.84NFLANAPQAGSKK827 pKa = 7.76SQRR830 pKa = 11.84AKK832 pKa = 10.94YY833 pKa = 8.02GTAGYY838 pKa = 9.76GVEE841 pKa = 4.38ARR843 pKa = 11.84GPTPEE848 pKa = 3.68EE849 pKa = 3.66AQRR852 pKa = 11.84EE853 pKa = 4.02KK854 pKa = 10.55DD855 pKa = 3.34TRR857 pKa = 11.84ISKK860 pKa = 10.43KK861 pKa = 9.06MEE863 pKa = 3.68TMGIYY868 pKa = 10.18FATPEE873 pKa = 3.79WVALNGHH880 pKa = 7.16RR881 pKa = 11.84GPSPGQVKK889 pKa = 9.23YY890 pKa = 9.19WQNKK894 pKa = 8.73RR895 pKa = 11.84EE896 pKa = 4.11IPDD899 pKa = 3.84PNEE902 pKa = 4.54DD903 pKa = 3.6YY904 pKa = 11.22LDD906 pKa = 3.79YY907 pKa = 11.55VHH909 pKa = 7.37AEE911 pKa = 3.96KK912 pKa = 10.86SRR914 pKa = 11.84LASEE918 pKa = 4.11EE919 pKa = 4.0QILRR923 pKa = 11.84AATSIYY929 pKa = 9.8GAPGQAEE936 pKa = 4.26PPQAFIDD943 pKa = 4.24EE944 pKa = 4.38VAKK947 pKa = 10.82VYY949 pKa = 10.07EE950 pKa = 4.43INHH953 pKa = 5.42GRR955 pKa = 11.84GPNQEE960 pKa = 3.86QMKK963 pKa = 10.54DD964 pKa = 3.44LLLTAMEE971 pKa = 4.32MKK973 pKa = 10.22HH974 pKa = 5.91RR975 pKa = 11.84NPRR978 pKa = 11.84RR979 pKa = 11.84ALPKK983 pKa = 10.15PKK985 pKa = 9.54PKK987 pKa = 10.28PNAPTQRR994 pKa = 11.84PPGRR998 pKa = 11.84LGRR1001 pKa = 11.84WIRR1004 pKa = 11.84TVSDD1008 pKa = 3.49EE1009 pKa = 4.72DD1010 pKa = 4.27LEE1012 pKa = 4.35
Molecular weight: 109.72 kDa
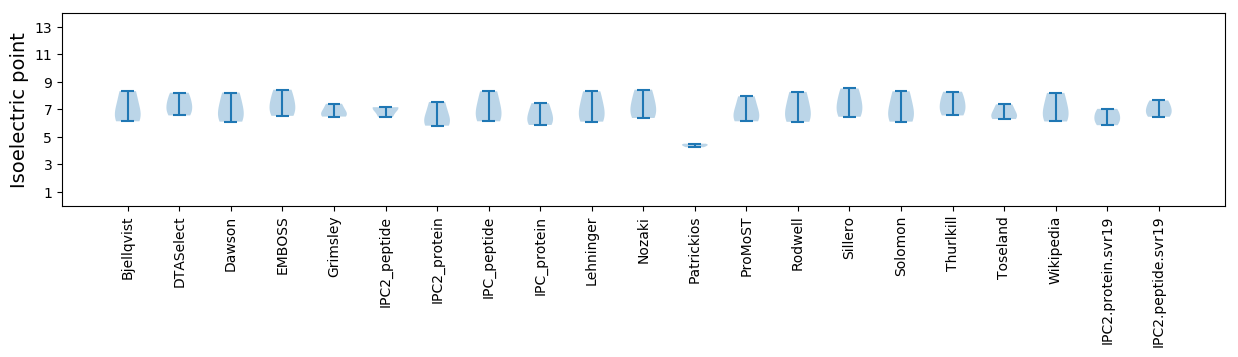
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P0C751|VP5_IBDVB Protein VP5 OS=Avian infectious bursal disease virus (strain Chicken/Cuba/Soroa/1998) OX=645118 GN=VP5 PE=3 SV=1
MM1 pKa = 7.68SDD3 pKa = 3.42VFNSPQARR11 pKa = 11.84STISAAFGIKK21 pKa = 8.03PTAGQDD27 pKa = 3.57VEE29 pKa = 4.51EE30 pKa = 4.15LLIPKK35 pKa = 9.41VWVPPEE41 pKa = 4.66DD42 pKa = 3.98PLASPSRR49 pKa = 11.84LAKK52 pKa = 10.1FLRR55 pKa = 11.84EE56 pKa = 3.65NGYY59 pKa = 10.25KK60 pKa = 9.98VLQPRR65 pKa = 11.84SLPEE69 pKa = 3.8NEE71 pKa = 4.84EE72 pKa = 4.32YY73 pKa = 9.31EE74 pKa = 4.26TDD76 pKa = 4.37QILPDD81 pKa = 4.33LAWMRR86 pKa = 11.84QIEE89 pKa = 4.46GAVLKK94 pKa = 8.25PTLSLPIGDD103 pKa = 3.76QEE105 pKa = 4.53YY106 pKa = 10.05FPKK109 pKa = 10.53YY110 pKa = 10.12YY111 pKa = 7.96PTHH114 pKa = 6.68RR115 pKa = 11.84PSKK118 pKa = 8.84EE119 pKa = 3.82KK120 pKa = 10.69PNAYY124 pKa = 9.47PPDD127 pKa = 3.66IALLKK132 pKa = 10.02QMIYY136 pKa = 10.94LFLQVPEE143 pKa = 4.35ANEE146 pKa = 3.96GLKK149 pKa = 10.98DD150 pKa = 3.89EE151 pKa = 4.54VTLLTQNIRR160 pKa = 11.84DD161 pKa = 3.64KK162 pKa = 11.21AYY164 pKa = 10.57GSGTYY169 pKa = 9.05MGQANRR175 pKa = 11.84LVAMKK180 pKa = 10.14EE181 pKa = 4.05VATGRR186 pKa = 11.84NPNKK190 pKa = 10.5DD191 pKa = 3.73PLKK194 pKa = 10.6LGYY197 pKa = 8.52TFEE200 pKa = 6.44SIAQLLDD207 pKa = 2.76ITLPVGPPGEE217 pKa = 4.58DD218 pKa = 3.24DD219 pKa = 4.36KK220 pKa = 11.71PWVPLTRR227 pKa = 11.84VPSRR231 pKa = 11.84MLVLTGDD238 pKa = 3.12VDD240 pKa = 4.05GDD242 pKa = 4.15FEE244 pKa = 5.23VEE246 pKa = 4.76DD247 pKa = 4.24YY248 pKa = 11.13LPKK251 pKa = 10.38INLKK255 pKa = 10.38SSSGLPYY262 pKa = 10.07VGRR265 pKa = 11.84TKK267 pKa = 11.16GEE269 pKa = 4.36TIGEE273 pKa = 4.19MIAISNQFLRR283 pKa = 11.84EE284 pKa = 3.93LSTLLKK290 pKa = 10.53QGAGTKK296 pKa = 10.27GSNKK300 pKa = 9.63KK301 pKa = 10.01KK302 pKa = 9.94LLSMLSDD309 pKa = 3.42YY310 pKa = 10.62WYY312 pKa = 10.51LSCGLLFPKK321 pKa = 10.44AEE323 pKa = 4.52RR324 pKa = 11.84YY325 pKa = 8.79DD326 pKa = 3.43KK327 pKa = 10.66STWLTKK333 pKa = 9.37TRR335 pKa = 11.84NIWSAPSPTHH345 pKa = 7.36LMISMITWPVMSNSPNNVLNIEE367 pKa = 4.55GCPSLYY373 pKa = 10.44KK374 pKa = 10.41FNPFRR379 pKa = 11.84GGLNRR384 pKa = 11.84IVEE387 pKa = 4.74WILAPEE393 pKa = 4.13EE394 pKa = 4.1PKK396 pKa = 11.08ALVYY400 pKa = 10.88ADD402 pKa = 4.13NIYY405 pKa = 10.24IVHH408 pKa = 6.42SNTWYY413 pKa = 10.94SIDD416 pKa = 4.29LEE418 pKa = 4.4KK419 pKa = 11.59GEE421 pKa = 4.61ANCTRR426 pKa = 11.84QHH428 pKa = 5.49MQAAMYY434 pKa = 10.0YY435 pKa = 8.69ILTRR439 pKa = 11.84GWSDD443 pKa = 3.09NGDD446 pKa = 3.46PMFNQTWATFAMNIAPALVVDD467 pKa = 4.6SSCLIMNLQIKK478 pKa = 9.15TYY480 pKa = 10.14GQGSGNAATFINNHH494 pKa = 5.92LLSTLVLDD502 pKa = 3.48QWNLMRR508 pKa = 11.84QPRR511 pKa = 11.84PDD513 pKa = 3.38SEE515 pKa = 4.2EE516 pKa = 3.85FKK518 pKa = 11.08SIEE521 pKa = 4.4DD522 pKa = 3.33KK523 pKa = 11.34LGINFKK529 pKa = 10.15IEE531 pKa = 3.77RR532 pKa = 11.84SIDD535 pKa = 3.93DD536 pKa = 2.97IRR538 pKa = 11.84GKK540 pKa = 10.39LRR542 pKa = 11.84QLVLLAQPGYY552 pKa = 10.59LSGGVEE558 pKa = 4.54PEE560 pKa = 3.89QSSPTVEE567 pKa = 4.37LDD569 pKa = 3.44LLGWSATYY577 pKa = 10.83SKK579 pKa = 11.21DD580 pKa = 3.02LGIYY584 pKa = 10.27VPVLDD589 pKa = 5.2KK590 pKa = 11.27EE591 pKa = 4.59RR592 pKa = 11.84LFCSAAYY599 pKa = 9.04PKK601 pKa = 10.69GVEE604 pKa = 4.06NKK606 pKa = 9.63SLKK609 pKa = 10.27SKK611 pKa = 10.93VGIEE615 pKa = 3.75QAYY618 pKa = 9.5KK619 pKa = 8.92VVRR622 pKa = 11.84YY623 pKa = 7.56EE624 pKa = 3.74ALRR627 pKa = 11.84LVGGWNYY634 pKa = 10.51PLLNKK639 pKa = 9.72ACKK642 pKa = 9.88NNAGAARR649 pKa = 11.84RR650 pKa = 11.84HH651 pKa = 5.68LEE653 pKa = 4.04AKK655 pKa = 10.1GFPLDD660 pKa = 3.58EE661 pKa = 5.47FLAEE665 pKa = 3.87WSEE668 pKa = 3.75LSEE671 pKa = 4.25FGEE674 pKa = 4.21AFEE677 pKa = 4.89GFNIKK682 pKa = 8.97LTVTSEE688 pKa = 4.12SLAEE692 pKa = 4.17LNKK695 pKa = 9.9PVPPKK700 pKa = 9.86PPNVNRR706 pKa = 11.84PVNTGGLKK714 pKa = 10.34AVSNALKK721 pKa = 9.43TGRR724 pKa = 11.84YY725 pKa = 8.86RR726 pKa = 11.84NEE728 pKa = 3.92AGLSGLVLLATARR741 pKa = 11.84SRR743 pKa = 11.84LQDD746 pKa = 3.35AVKK749 pKa = 10.7AKK751 pKa = 10.94AEE753 pKa = 3.94AEE755 pKa = 4.15KK756 pKa = 10.69LHH758 pKa = 6.52KK759 pKa = 10.57SKK761 pKa = 10.83PDD763 pKa = 3.84DD764 pKa = 4.69PDD766 pKa = 5.73ADD768 pKa = 3.12WFEE771 pKa = 4.42RR772 pKa = 11.84SEE774 pKa = 4.24TLSDD778 pKa = 3.95LLEE781 pKa = 4.43KK782 pKa = 10.72ADD784 pKa = 3.58IASKK788 pKa = 9.84VAHH791 pKa = 6.17SALVEE796 pKa = 4.02TSDD799 pKa = 4.36ALEE802 pKa = 4.48AVQSTSVYY810 pKa = 7.57TPKK813 pKa = 10.61YY814 pKa = 9.41PEE816 pKa = 3.92VKK818 pKa = 10.55NPQTASNPVVGLHH831 pKa = 6.53LPAKK835 pKa = 10.0RR836 pKa = 11.84ATGVQAALLGAGTSRR851 pKa = 11.84PMGMEE856 pKa = 3.61APTRR860 pKa = 11.84SKK862 pKa = 11.17NAVKK866 pKa = 9.47MAKK869 pKa = 9.48RR870 pKa = 11.84RR871 pKa = 11.84QRR873 pKa = 11.84QKK875 pKa = 10.89EE876 pKa = 4.04SRR878 pKa = 11.84QQ879 pKa = 3.42
MM1 pKa = 7.68SDD3 pKa = 3.42VFNSPQARR11 pKa = 11.84STISAAFGIKK21 pKa = 8.03PTAGQDD27 pKa = 3.57VEE29 pKa = 4.51EE30 pKa = 4.15LLIPKK35 pKa = 9.41VWVPPEE41 pKa = 4.66DD42 pKa = 3.98PLASPSRR49 pKa = 11.84LAKK52 pKa = 10.1FLRR55 pKa = 11.84EE56 pKa = 3.65NGYY59 pKa = 10.25KK60 pKa = 9.98VLQPRR65 pKa = 11.84SLPEE69 pKa = 3.8NEE71 pKa = 4.84EE72 pKa = 4.32YY73 pKa = 9.31EE74 pKa = 4.26TDD76 pKa = 4.37QILPDD81 pKa = 4.33LAWMRR86 pKa = 11.84QIEE89 pKa = 4.46GAVLKK94 pKa = 8.25PTLSLPIGDD103 pKa = 3.76QEE105 pKa = 4.53YY106 pKa = 10.05FPKK109 pKa = 10.53YY110 pKa = 10.12YY111 pKa = 7.96PTHH114 pKa = 6.68RR115 pKa = 11.84PSKK118 pKa = 8.84EE119 pKa = 3.82KK120 pKa = 10.69PNAYY124 pKa = 9.47PPDD127 pKa = 3.66IALLKK132 pKa = 10.02QMIYY136 pKa = 10.94LFLQVPEE143 pKa = 4.35ANEE146 pKa = 3.96GLKK149 pKa = 10.98DD150 pKa = 3.89EE151 pKa = 4.54VTLLTQNIRR160 pKa = 11.84DD161 pKa = 3.64KK162 pKa = 11.21AYY164 pKa = 10.57GSGTYY169 pKa = 9.05MGQANRR175 pKa = 11.84LVAMKK180 pKa = 10.14EE181 pKa = 4.05VATGRR186 pKa = 11.84NPNKK190 pKa = 10.5DD191 pKa = 3.73PLKK194 pKa = 10.6LGYY197 pKa = 8.52TFEE200 pKa = 6.44SIAQLLDD207 pKa = 2.76ITLPVGPPGEE217 pKa = 4.58DD218 pKa = 3.24DD219 pKa = 4.36KK220 pKa = 11.71PWVPLTRR227 pKa = 11.84VPSRR231 pKa = 11.84MLVLTGDD238 pKa = 3.12VDD240 pKa = 4.05GDD242 pKa = 4.15FEE244 pKa = 5.23VEE246 pKa = 4.76DD247 pKa = 4.24YY248 pKa = 11.13LPKK251 pKa = 10.38INLKK255 pKa = 10.38SSSGLPYY262 pKa = 10.07VGRR265 pKa = 11.84TKK267 pKa = 11.16GEE269 pKa = 4.36TIGEE273 pKa = 4.19MIAISNQFLRR283 pKa = 11.84EE284 pKa = 3.93LSTLLKK290 pKa = 10.53QGAGTKK296 pKa = 10.27GSNKK300 pKa = 9.63KK301 pKa = 10.01KK302 pKa = 9.94LLSMLSDD309 pKa = 3.42YY310 pKa = 10.62WYY312 pKa = 10.51LSCGLLFPKK321 pKa = 10.44AEE323 pKa = 4.52RR324 pKa = 11.84YY325 pKa = 8.79DD326 pKa = 3.43KK327 pKa = 10.66STWLTKK333 pKa = 9.37TRR335 pKa = 11.84NIWSAPSPTHH345 pKa = 7.36LMISMITWPVMSNSPNNVLNIEE367 pKa = 4.55GCPSLYY373 pKa = 10.44KK374 pKa = 10.41FNPFRR379 pKa = 11.84GGLNRR384 pKa = 11.84IVEE387 pKa = 4.74WILAPEE393 pKa = 4.13EE394 pKa = 4.1PKK396 pKa = 11.08ALVYY400 pKa = 10.88ADD402 pKa = 4.13NIYY405 pKa = 10.24IVHH408 pKa = 6.42SNTWYY413 pKa = 10.94SIDD416 pKa = 4.29LEE418 pKa = 4.4KK419 pKa = 11.59GEE421 pKa = 4.61ANCTRR426 pKa = 11.84QHH428 pKa = 5.49MQAAMYY434 pKa = 10.0YY435 pKa = 8.69ILTRR439 pKa = 11.84GWSDD443 pKa = 3.09NGDD446 pKa = 3.46PMFNQTWATFAMNIAPALVVDD467 pKa = 4.6SSCLIMNLQIKK478 pKa = 9.15TYY480 pKa = 10.14GQGSGNAATFINNHH494 pKa = 5.92LLSTLVLDD502 pKa = 3.48QWNLMRR508 pKa = 11.84QPRR511 pKa = 11.84PDD513 pKa = 3.38SEE515 pKa = 4.2EE516 pKa = 3.85FKK518 pKa = 11.08SIEE521 pKa = 4.4DD522 pKa = 3.33KK523 pKa = 11.34LGINFKK529 pKa = 10.15IEE531 pKa = 3.77RR532 pKa = 11.84SIDD535 pKa = 3.93DD536 pKa = 2.97IRR538 pKa = 11.84GKK540 pKa = 10.39LRR542 pKa = 11.84QLVLLAQPGYY552 pKa = 10.59LSGGVEE558 pKa = 4.54PEE560 pKa = 3.89QSSPTVEE567 pKa = 4.37LDD569 pKa = 3.44LLGWSATYY577 pKa = 10.83SKK579 pKa = 11.21DD580 pKa = 3.02LGIYY584 pKa = 10.27VPVLDD589 pKa = 5.2KK590 pKa = 11.27EE591 pKa = 4.59RR592 pKa = 11.84LFCSAAYY599 pKa = 9.04PKK601 pKa = 10.69GVEE604 pKa = 4.06NKK606 pKa = 9.63SLKK609 pKa = 10.27SKK611 pKa = 10.93VGIEE615 pKa = 3.75QAYY618 pKa = 9.5KK619 pKa = 8.92VVRR622 pKa = 11.84YY623 pKa = 7.56EE624 pKa = 3.74ALRR627 pKa = 11.84LVGGWNYY634 pKa = 10.51PLLNKK639 pKa = 9.72ACKK642 pKa = 9.88NNAGAARR649 pKa = 11.84RR650 pKa = 11.84HH651 pKa = 5.68LEE653 pKa = 4.04AKK655 pKa = 10.1GFPLDD660 pKa = 3.58EE661 pKa = 5.47FLAEE665 pKa = 3.87WSEE668 pKa = 3.75LSEE671 pKa = 4.25FGEE674 pKa = 4.21AFEE677 pKa = 4.89GFNIKK682 pKa = 8.97LTVTSEE688 pKa = 4.12SLAEE692 pKa = 4.17LNKK695 pKa = 9.9PVPPKK700 pKa = 9.86PPNVNRR706 pKa = 11.84PVNTGGLKK714 pKa = 10.34AVSNALKK721 pKa = 9.43TGRR724 pKa = 11.84YY725 pKa = 8.86RR726 pKa = 11.84NEE728 pKa = 3.92AGLSGLVLLATARR741 pKa = 11.84SRR743 pKa = 11.84LQDD746 pKa = 3.35AVKK749 pKa = 10.7AKK751 pKa = 10.94AEE753 pKa = 3.94AEE755 pKa = 4.15KK756 pKa = 10.69LHH758 pKa = 6.52KK759 pKa = 10.57SKK761 pKa = 10.83PDD763 pKa = 3.84DD764 pKa = 4.69PDD766 pKa = 5.73ADD768 pKa = 3.12WFEE771 pKa = 4.42RR772 pKa = 11.84SEE774 pKa = 4.24TLSDD778 pKa = 3.95LLEE781 pKa = 4.43KK782 pKa = 10.72ADD784 pKa = 3.58IASKK788 pKa = 9.84VAHH791 pKa = 6.17SALVEE796 pKa = 4.02TSDD799 pKa = 4.36ALEE802 pKa = 4.48AVQSTSVYY810 pKa = 7.57TPKK813 pKa = 10.61YY814 pKa = 9.41PEE816 pKa = 3.92VKK818 pKa = 10.55NPQTASNPVVGLHH831 pKa = 6.53LPAKK835 pKa = 10.0RR836 pKa = 11.84ATGVQAALLGAGTSRR851 pKa = 11.84PMGMEE856 pKa = 3.61APTRR860 pKa = 11.84SKK862 pKa = 11.17NAVKK866 pKa = 9.47MAKK869 pKa = 9.48RR870 pKa = 11.84RR871 pKa = 11.84QRR873 pKa = 11.84QKK875 pKa = 10.89EE876 pKa = 4.04SRR878 pKa = 11.84QQ879 pKa = 3.42
Molecular weight: 97.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2036 |
145 |
1012 |
678.7 |
74.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.644 ± 1.039 | 0.835 ± 0.926 |
5.157 ± 0.734 | 5.796 ± 0.64 |
2.898 ± 0.433 | 7.417 ± 0.814 |
1.719 ± 1.204 | 4.568 ± 0.96 |
5.059 ± 1.304 | 9.823 ± 0.948 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.21 ± 0.352 | 4.961 ± 0.23 |
7.318 ± 0.062 | 3.487 ± 0.485 |
5.648 ± 0.985 | 6.974 ± 0.737 |
6.139 ± 0.774 | 6.532 ± 0.565 |
1.473 ± 0.556 | 3.34 ± 0.769 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
