
Acholeplasma oculi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Acholeplasmatales; Acholeplasmataceae; Acholeplasma
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
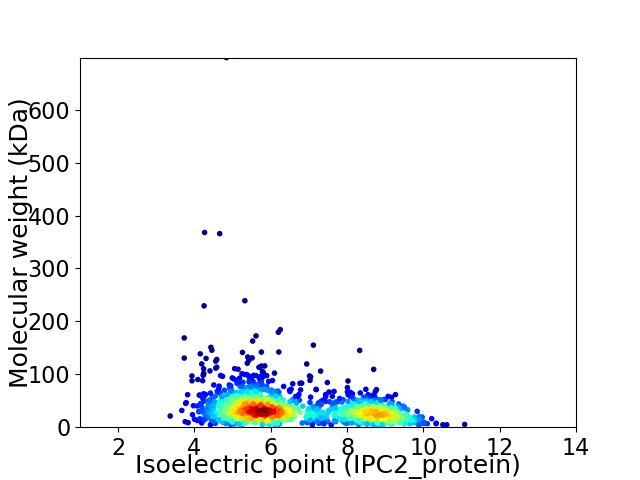
Virtual 2D-PAGE plot for 1468 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
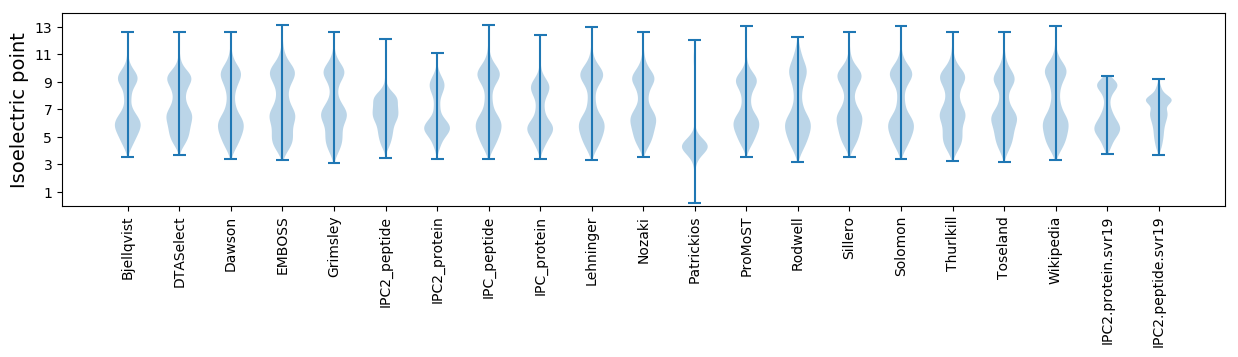
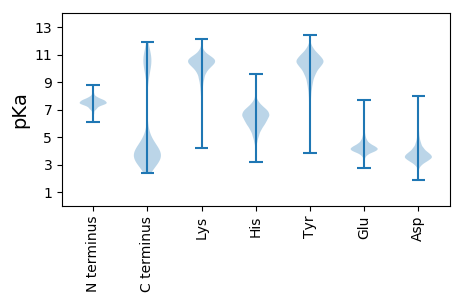
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A061AGP1|A0A061AGP1_9MOLU Uncharacterized protein OS=Acholeplasma oculi OX=35623 GN=Aocu_00540 PE=4 SV=1
MM1 pKa = 7.29KK2 pKa = 10.14KK3 pKa = 10.42VYY5 pKa = 8.72TALFILAAVFILNACGSNEE24 pKa = 3.72LVIYY28 pKa = 8.67MPKK31 pKa = 10.23EE32 pKa = 4.01YY33 pKa = 9.81ISEE36 pKa = 4.17DD37 pKa = 3.15VVMAFEE43 pKa = 4.35AEE45 pKa = 4.12KK46 pKa = 10.64GIKK49 pKa = 9.86VSLRR53 pKa = 11.84YY54 pKa = 9.28FDD56 pKa = 4.35SNEE59 pKa = 3.83VLLTNAKK66 pKa = 9.77VNSYY70 pKa = 11.24DD71 pKa = 4.44LIIPSDD77 pKa = 3.78YY78 pKa = 10.95AIEE81 pKa = 4.04EE82 pKa = 4.29LATEE86 pKa = 4.74GFLKK90 pKa = 10.78KK91 pKa = 10.43LDD93 pKa = 3.35WEE95 pKa = 4.91KK96 pKa = 10.91IDD98 pKa = 4.46FEE100 pKa = 4.42SSEE103 pKa = 4.26MNEE106 pKa = 4.22SLVTTLNQLEE116 pKa = 4.26SDD118 pKa = 3.57GFDD121 pKa = 3.08FLEE124 pKa = 4.3YY125 pKa = 10.79SMPYY129 pKa = 9.23FWGTVGLIYY138 pKa = 10.92NNTIEE143 pKa = 4.45GLEE146 pKa = 4.09SDD148 pKa = 4.5VEE150 pKa = 4.28TLEE153 pKa = 4.0WAILNEE159 pKa = 4.31SKK161 pKa = 10.33YY162 pKa = 11.36SKK164 pKa = 9.99MIYY167 pKa = 10.01DD168 pKa = 4.08SSRR171 pKa = 11.84DD172 pKa = 3.25AFMAGLYY179 pKa = 11.02ANDD182 pKa = 3.7LRR184 pKa = 11.84MANPTQTNINTAKK197 pKa = 10.77DD198 pKa = 3.65FLIDD202 pKa = 4.17ANSKK206 pKa = 10.47SNASIKK212 pKa = 10.28SDD214 pKa = 3.86EE215 pKa = 4.57ILTEE219 pKa = 4.66AIGGQTPYY227 pKa = 10.68DD228 pKa = 3.46VAMVYY233 pKa = 10.49SGDD236 pKa = 3.39AVYY239 pKa = 10.52ILQEE243 pKa = 3.67TDD245 pKa = 2.69NYY247 pKa = 10.86SFYY250 pKa = 11.14VPEE253 pKa = 4.46WSNVWIDD260 pKa = 3.95GFVIPNNTKK269 pKa = 10.29HH270 pKa = 6.55EE271 pKa = 4.07DD272 pKa = 2.97WAYY275 pKa = 10.36EE276 pKa = 4.1FINFMSSYY284 pKa = 9.76EE285 pKa = 3.84ASLEE289 pKa = 3.74NTYY292 pKa = 11.74AMGYY296 pKa = 7.43TPVRR300 pKa = 11.84QDD302 pKa = 3.32VLEE305 pKa = 4.28EE306 pKa = 3.96LLADD310 pKa = 5.52EE311 pKa = 4.68EE312 pKa = 4.62FAWDD316 pKa = 4.23DD317 pKa = 4.04RR318 pKa = 11.84ISYY321 pKa = 10.44AFTVPVVNFEE331 pKa = 4.19FYY333 pKa = 10.47RR334 pKa = 11.84YY335 pKa = 9.54NATLKK340 pKa = 11.2SMIDD344 pKa = 3.73DD345 pKa = 3.24AWEE348 pKa = 3.77LVILSNN354 pKa = 5.04
MM1 pKa = 7.29KK2 pKa = 10.14KK3 pKa = 10.42VYY5 pKa = 8.72TALFILAAVFILNACGSNEE24 pKa = 3.72LVIYY28 pKa = 8.67MPKK31 pKa = 10.23EE32 pKa = 4.01YY33 pKa = 9.81ISEE36 pKa = 4.17DD37 pKa = 3.15VVMAFEE43 pKa = 4.35AEE45 pKa = 4.12KK46 pKa = 10.64GIKK49 pKa = 9.86VSLRR53 pKa = 11.84YY54 pKa = 9.28FDD56 pKa = 4.35SNEE59 pKa = 3.83VLLTNAKK66 pKa = 9.77VNSYY70 pKa = 11.24DD71 pKa = 4.44LIIPSDD77 pKa = 3.78YY78 pKa = 10.95AIEE81 pKa = 4.04EE82 pKa = 4.29LATEE86 pKa = 4.74GFLKK90 pKa = 10.78KK91 pKa = 10.43LDD93 pKa = 3.35WEE95 pKa = 4.91KK96 pKa = 10.91IDD98 pKa = 4.46FEE100 pKa = 4.42SSEE103 pKa = 4.26MNEE106 pKa = 4.22SLVTTLNQLEE116 pKa = 4.26SDD118 pKa = 3.57GFDD121 pKa = 3.08FLEE124 pKa = 4.3YY125 pKa = 10.79SMPYY129 pKa = 9.23FWGTVGLIYY138 pKa = 10.92NNTIEE143 pKa = 4.45GLEE146 pKa = 4.09SDD148 pKa = 4.5VEE150 pKa = 4.28TLEE153 pKa = 4.0WAILNEE159 pKa = 4.31SKK161 pKa = 10.33YY162 pKa = 11.36SKK164 pKa = 9.99MIYY167 pKa = 10.01DD168 pKa = 4.08SSRR171 pKa = 11.84DD172 pKa = 3.25AFMAGLYY179 pKa = 11.02ANDD182 pKa = 3.7LRR184 pKa = 11.84MANPTQTNINTAKK197 pKa = 10.77DD198 pKa = 3.65FLIDD202 pKa = 4.17ANSKK206 pKa = 10.47SNASIKK212 pKa = 10.28SDD214 pKa = 3.86EE215 pKa = 4.57ILTEE219 pKa = 4.66AIGGQTPYY227 pKa = 10.68DD228 pKa = 3.46VAMVYY233 pKa = 10.49SGDD236 pKa = 3.39AVYY239 pKa = 10.52ILQEE243 pKa = 3.67TDD245 pKa = 2.69NYY247 pKa = 10.86SFYY250 pKa = 11.14VPEE253 pKa = 4.46WSNVWIDD260 pKa = 3.95GFVIPNNTKK269 pKa = 10.29HH270 pKa = 6.55EE271 pKa = 4.07DD272 pKa = 2.97WAYY275 pKa = 10.36EE276 pKa = 4.1FINFMSSYY284 pKa = 9.76EE285 pKa = 3.84ASLEE289 pKa = 3.74NTYY292 pKa = 11.74AMGYY296 pKa = 7.43TPVRR300 pKa = 11.84QDD302 pKa = 3.32VLEE305 pKa = 4.28EE306 pKa = 3.96LLADD310 pKa = 5.52EE311 pKa = 4.68EE312 pKa = 4.62FAWDD316 pKa = 4.23DD317 pKa = 4.04RR318 pKa = 11.84ISYY321 pKa = 10.44AFTVPVVNFEE331 pKa = 4.19FYY333 pKa = 10.47RR334 pKa = 11.84YY335 pKa = 9.54NATLKK340 pKa = 11.2SMIDD344 pKa = 3.73DD345 pKa = 3.24AWEE348 pKa = 3.77LVILSNN354 pKa = 5.04
Molecular weight: 40.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A061AF12|A0A061AF12_9MOLU Glutamine--tRNA ligase OS=Acholeplasma oculi OX=35623 GN=glnS PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.95QPSKK9 pKa = 9.64IKK11 pKa = 10.09HH12 pKa = 4.75QRR14 pKa = 11.84THH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84HH40 pKa = 4.39VLTVV44 pKa = 3.21
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.95QPSKK9 pKa = 9.64IKK11 pKa = 10.09HH12 pKa = 4.75QRR14 pKa = 11.84THH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84HH40 pKa = 4.39VLTVV44 pKa = 3.21
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
484631 |
37 |
6297 |
330.1 |
37.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.322 ± 0.059 | 0.408 ± 0.016 |
5.848 ± 0.048 | 6.491 ± 0.06 |
5.016 ± 0.048 | 5.879 ± 0.08 |
2.115 ± 0.033 | 9.109 ± 0.068 |
7.851 ± 0.102 | 10.193 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.587 ± 0.03 | 5.405 ± 0.051 |
3.064 ± 0.033 | 3.398 ± 0.038 |
3.36 ± 0.044 | 6.26 ± 0.05 |
5.646 ± 0.098 | 6.596 ± 0.067 |
0.752 ± 0.02 | 4.701 ± 0.062 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
