
Japanese holly fern mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Mayoviridae; Pteridovirus; Japanese holly fern mottle pteridovirus
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
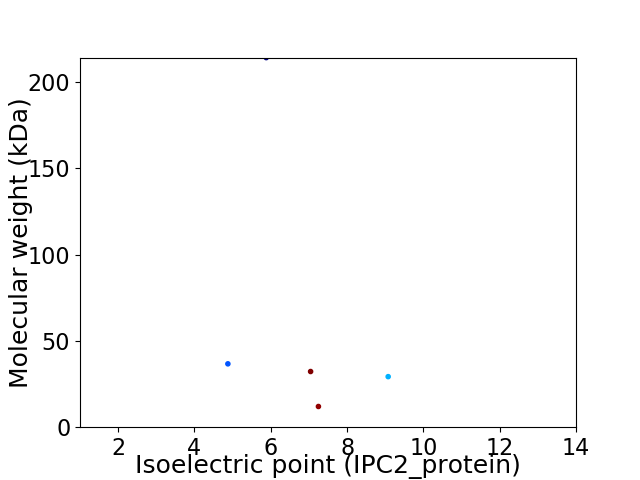
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7T4Z8|C7T4Z8_9VIRU p29 OS=Japanese holly fern mottle virus OX=659660 PE=4 SV=1
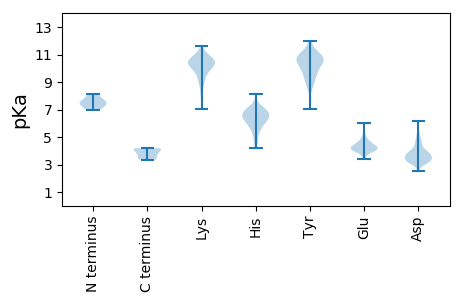
MM1 pKa = 7.51KK2 pKa = 10.1GASGLRR8 pKa = 11.84RR9 pKa = 11.84LGFRR13 pKa = 11.84AVVPAVVVGGAVAGTVVHH31 pKa = 6.84RR32 pKa = 11.84ALNNRR37 pKa = 11.84GGDD40 pKa = 3.89GEE42 pKa = 4.13NLRR45 pKa = 11.84DD46 pKa = 3.86LRR48 pKa = 11.84EE49 pKa = 3.99LRR51 pKa = 11.84PLEE54 pKa = 4.02VEE56 pKa = 4.11EE57 pKa = 4.27FTISSGDD64 pKa = 3.7SVAEE68 pKa = 4.36SIDD71 pKa = 3.4IDD73 pKa = 3.92SAAVNSAADD82 pKa = 4.19LVPFDD87 pKa = 4.82AVLEE91 pKa = 4.19PEE93 pKa = 4.59EE94 pKa = 4.69PVTLPVSNPRR104 pKa = 11.84RR105 pKa = 11.84IWSRR109 pKa = 11.84LFAAMVTLSVPLAGVFAVRR128 pKa = 11.84HH129 pKa = 4.21VTGQGSVSAGDD140 pKa = 3.93DD141 pKa = 3.57LRR143 pKa = 11.84PEE145 pKa = 4.15PFGPRR150 pKa = 11.84DD151 pKa = 3.41VPRR154 pKa = 11.84PDD156 pKa = 4.7EE157 pKa = 4.63IPPHH161 pKa = 5.9AAVEE165 pKa = 4.21EE166 pKa = 4.44GDD168 pKa = 4.01PVACLPEE175 pKa = 4.24DD176 pKa = 2.96SRR178 pKa = 11.84RR179 pKa = 11.84FVKK182 pKa = 10.53RR183 pKa = 11.84CLDD186 pKa = 3.27HH187 pKa = 6.83LAYY190 pKa = 10.43FEE192 pKa = 4.59LVGVCPFCEE201 pKa = 3.81RR202 pKa = 11.84DD203 pKa = 3.51GLKK206 pKa = 10.55SISFEE211 pKa = 3.46AVVFEE216 pKa = 4.28EE217 pKa = 4.55VPPLFVGKK225 pKa = 10.66FEE227 pKa = 6.05LIPSAVFDD235 pKa = 3.58PHH237 pKa = 7.07FWEE240 pKa = 4.42PLCYY244 pKa = 9.09YY245 pKa = 10.31TLFQDD250 pKa = 4.87MLVCKK255 pKa = 10.13CGKK258 pKa = 8.54HH259 pKa = 6.28DD260 pKa = 3.94RR261 pKa = 11.84YY262 pKa = 10.34HH263 pKa = 5.63VVKK266 pKa = 9.79IYY268 pKa = 10.6PVMDD272 pKa = 3.23VHH274 pKa = 6.98PLNTVYY280 pKa = 10.46MLFIVKK286 pKa = 10.04EE287 pKa = 4.13GFDD290 pKa = 3.66VDD292 pKa = 3.58RR293 pKa = 11.84GFPVMSSYY301 pKa = 10.98GYY303 pKa = 8.84HH304 pKa = 6.11VGWAFEE310 pKa = 4.1PLTIPLEE317 pKa = 4.41PFDD320 pKa = 3.98GTEE323 pKa = 4.02PLPLHH328 pKa = 6.56NEE330 pKa = 4.01TGRR333 pKa = 3.54
MM1 pKa = 7.51KK2 pKa = 10.1GASGLRR8 pKa = 11.84RR9 pKa = 11.84LGFRR13 pKa = 11.84AVVPAVVVGGAVAGTVVHH31 pKa = 6.84RR32 pKa = 11.84ALNNRR37 pKa = 11.84GGDD40 pKa = 3.89GEE42 pKa = 4.13NLRR45 pKa = 11.84DD46 pKa = 3.86LRR48 pKa = 11.84EE49 pKa = 3.99LRR51 pKa = 11.84PLEE54 pKa = 4.02VEE56 pKa = 4.11EE57 pKa = 4.27FTISSGDD64 pKa = 3.7SVAEE68 pKa = 4.36SIDD71 pKa = 3.4IDD73 pKa = 3.92SAAVNSAADD82 pKa = 4.19LVPFDD87 pKa = 4.82AVLEE91 pKa = 4.19PEE93 pKa = 4.59EE94 pKa = 4.69PVTLPVSNPRR104 pKa = 11.84RR105 pKa = 11.84IWSRR109 pKa = 11.84LFAAMVTLSVPLAGVFAVRR128 pKa = 11.84HH129 pKa = 4.21VTGQGSVSAGDD140 pKa = 3.93DD141 pKa = 3.57LRR143 pKa = 11.84PEE145 pKa = 4.15PFGPRR150 pKa = 11.84DD151 pKa = 3.41VPRR154 pKa = 11.84PDD156 pKa = 4.7EE157 pKa = 4.63IPPHH161 pKa = 5.9AAVEE165 pKa = 4.21EE166 pKa = 4.44GDD168 pKa = 4.01PVACLPEE175 pKa = 4.24DD176 pKa = 2.96SRR178 pKa = 11.84RR179 pKa = 11.84FVKK182 pKa = 10.53RR183 pKa = 11.84CLDD186 pKa = 3.27HH187 pKa = 6.83LAYY190 pKa = 10.43FEE192 pKa = 4.59LVGVCPFCEE201 pKa = 3.81RR202 pKa = 11.84DD203 pKa = 3.51GLKK206 pKa = 10.55SISFEE211 pKa = 3.46AVVFEE216 pKa = 4.28EE217 pKa = 4.55VPPLFVGKK225 pKa = 10.66FEE227 pKa = 6.05LIPSAVFDD235 pKa = 3.58PHH237 pKa = 7.07FWEE240 pKa = 4.42PLCYY244 pKa = 9.09YY245 pKa = 10.31TLFQDD250 pKa = 4.87MLVCKK255 pKa = 10.13CGKK258 pKa = 8.54HH259 pKa = 6.28DD260 pKa = 3.94RR261 pKa = 11.84YY262 pKa = 10.34HH263 pKa = 5.63VVKK266 pKa = 9.79IYY268 pKa = 10.6PVMDD272 pKa = 3.23VHH274 pKa = 6.98PLNTVYY280 pKa = 10.46MLFIVKK286 pKa = 10.04EE287 pKa = 4.13GFDD290 pKa = 3.66VDD292 pKa = 3.58RR293 pKa = 11.84GFPVMSSYY301 pKa = 10.98GYY303 pKa = 8.84HH304 pKa = 6.11VGWAFEE310 pKa = 4.1PLTIPLEE317 pKa = 4.41PFDD320 pKa = 3.98GTEE323 pKa = 4.02PLPLHH328 pKa = 6.56NEE330 pKa = 4.01TGRR333 pKa = 3.54
Molecular weight: 36.73 kDa
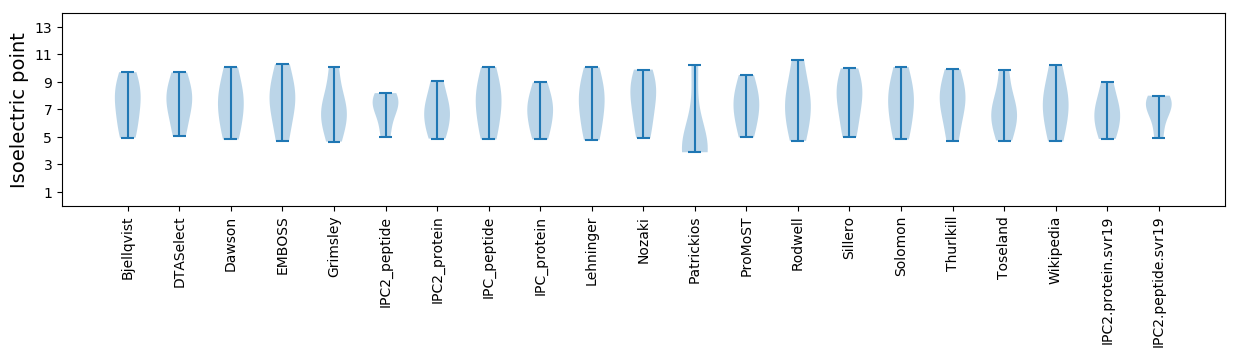
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7T4Z8|C7T4Z8_9VIRU p29 OS=Japanese holly fern mottle virus OX=659660 PE=4 SV=1
MM1 pKa = 7.55APKK4 pKa = 10.14ALPKK8 pKa = 10.31NVPKK12 pKa = 10.11EE13 pKa = 3.79YY14 pKa = 11.12AFIGPKK20 pKa = 9.88LWPKK24 pKa = 9.66LQEE27 pKa = 4.57AISYY31 pKa = 10.36GFDD34 pKa = 2.75IRR36 pKa = 11.84KK37 pKa = 9.68ARR39 pKa = 11.84DD40 pKa = 3.29DD41 pKa = 4.64GMTLSNLPFVEE52 pKa = 4.27KK53 pKa = 10.63EE54 pKa = 3.85KK55 pKa = 11.09VLNAFLKK62 pKa = 10.42SKK64 pKa = 10.12RR65 pKa = 11.84KK66 pKa = 9.65SGSSSSSTTGKK77 pKa = 10.18VPVRR81 pKa = 11.84DD82 pKa = 3.77VPEE85 pKa = 4.05SSVARR90 pKa = 11.84KK91 pKa = 8.08EE92 pKa = 4.08KK93 pKa = 10.02PRR95 pKa = 11.84KK96 pKa = 9.01QKK98 pKa = 8.91PTGVFMWEE106 pKa = 3.8PASLIGVDD114 pKa = 3.69GDD116 pKa = 3.89PGKK119 pKa = 10.41FAALPHH125 pKa = 6.24GKK127 pKa = 9.45SAVLLSFKK135 pKa = 10.56PPASPCMISKK145 pKa = 8.94MLIMLKK151 pKa = 10.14VDD153 pKa = 3.61ATADD157 pKa = 3.73LVVPYY162 pKa = 10.66AGITGIDD169 pKa = 3.45HH170 pKa = 7.21AGDD173 pKa = 3.77LAADD177 pKa = 4.64RR178 pKa = 11.84YY179 pKa = 11.11SVMVVNSRR187 pKa = 11.84YY188 pKa = 8.5PTMKK192 pKa = 10.54LLEE195 pKa = 4.79ADD197 pKa = 4.17DD198 pKa = 4.87KK199 pKa = 11.15ISYY202 pKa = 10.22RR203 pKa = 11.84FTFDD207 pKa = 3.51EE208 pKa = 4.9PVSSDD213 pKa = 3.02LFSRR217 pKa = 11.84IRR219 pKa = 11.84FIVTTANVAKK229 pKa = 9.27TKK231 pKa = 10.31SWLYY235 pKa = 7.68CQCWYY240 pKa = 10.72KK241 pKa = 11.03YY242 pKa = 10.07RR243 pKa = 11.84INKK246 pKa = 8.09TPVEE250 pKa = 3.97KK251 pKa = 10.34GGRR254 pKa = 11.84VMAVRR259 pKa = 11.84FDD261 pKa = 3.58AGAVV265 pKa = 3.35
MM1 pKa = 7.55APKK4 pKa = 10.14ALPKK8 pKa = 10.31NVPKK12 pKa = 10.11EE13 pKa = 3.79YY14 pKa = 11.12AFIGPKK20 pKa = 9.88LWPKK24 pKa = 9.66LQEE27 pKa = 4.57AISYY31 pKa = 10.36GFDD34 pKa = 2.75IRR36 pKa = 11.84KK37 pKa = 9.68ARR39 pKa = 11.84DD40 pKa = 3.29DD41 pKa = 4.64GMTLSNLPFVEE52 pKa = 4.27KK53 pKa = 10.63EE54 pKa = 3.85KK55 pKa = 11.09VLNAFLKK62 pKa = 10.42SKK64 pKa = 10.12RR65 pKa = 11.84KK66 pKa = 9.65SGSSSSSTTGKK77 pKa = 10.18VPVRR81 pKa = 11.84DD82 pKa = 3.77VPEE85 pKa = 4.05SSVARR90 pKa = 11.84KK91 pKa = 8.08EE92 pKa = 4.08KK93 pKa = 10.02PRR95 pKa = 11.84KK96 pKa = 9.01QKK98 pKa = 8.91PTGVFMWEE106 pKa = 3.8PASLIGVDD114 pKa = 3.69GDD116 pKa = 3.89PGKK119 pKa = 10.41FAALPHH125 pKa = 6.24GKK127 pKa = 9.45SAVLLSFKK135 pKa = 10.56PPASPCMISKK145 pKa = 8.94MLIMLKK151 pKa = 10.14VDD153 pKa = 3.61ATADD157 pKa = 3.73LVVPYY162 pKa = 10.66AGITGIDD169 pKa = 3.45HH170 pKa = 7.21AGDD173 pKa = 3.77LAADD177 pKa = 4.64RR178 pKa = 11.84YY179 pKa = 11.11SVMVVNSRR187 pKa = 11.84YY188 pKa = 8.5PTMKK192 pKa = 10.54LLEE195 pKa = 4.79ADD197 pKa = 4.17DD198 pKa = 4.87KK199 pKa = 11.15ISYY202 pKa = 10.22RR203 pKa = 11.84FTFDD207 pKa = 3.51EE208 pKa = 4.9PVSSDD213 pKa = 3.02LFSRR217 pKa = 11.84IRR219 pKa = 11.84FIVTTANVAKK229 pKa = 9.27TKK231 pKa = 10.31SWLYY235 pKa = 7.68CQCWYY240 pKa = 10.72KK241 pKa = 11.03YY242 pKa = 10.07RR243 pKa = 11.84INKK246 pKa = 8.09TPVEE250 pKa = 3.97KK251 pKa = 10.34GGRR254 pKa = 11.84VMAVRR259 pKa = 11.84FDD261 pKa = 3.58AGAVV265 pKa = 3.35
Molecular weight: 29.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2911 |
106 |
1909 |
582.2 |
64.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.561 ± 0.428 | 2.576 ± 0.461 |
6.699 ± 0.56 | 5.119 ± 0.517 |
5.29 ± 0.331 | 5.29 ± 0.831 |
2.336 ± 0.51 | 3.813 ± 0.592 |
5.496 ± 1.008 | 9.035 ± 0.592 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.37 ± 0.262 | 2.92 ± 0.809 |
5.737 ± 1.231 | 2.267 ± 0.474 |
5.806 ± 0.263 | 9.619 ± 1.008 |
4.706 ± 0.509 | 10.271 ± 0.692 |
1.099 ± 0.165 | 2.989 ± 0.223 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
