
Pseudobythopirellula maris
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Lacipirellulaceae; Pseudobythopirellula
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
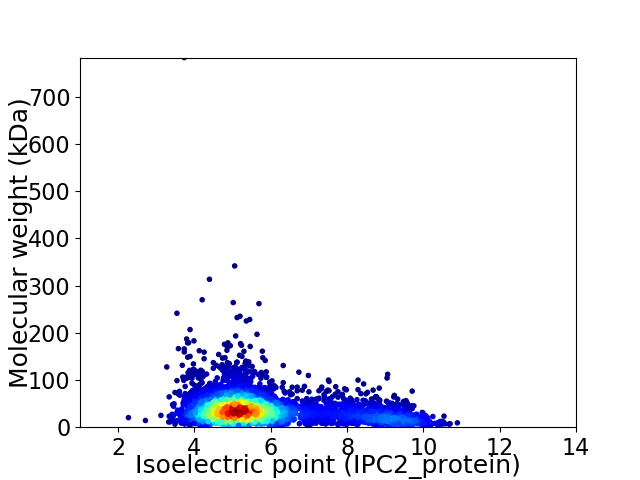
Virtual 2D-PAGE plot for 3893 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
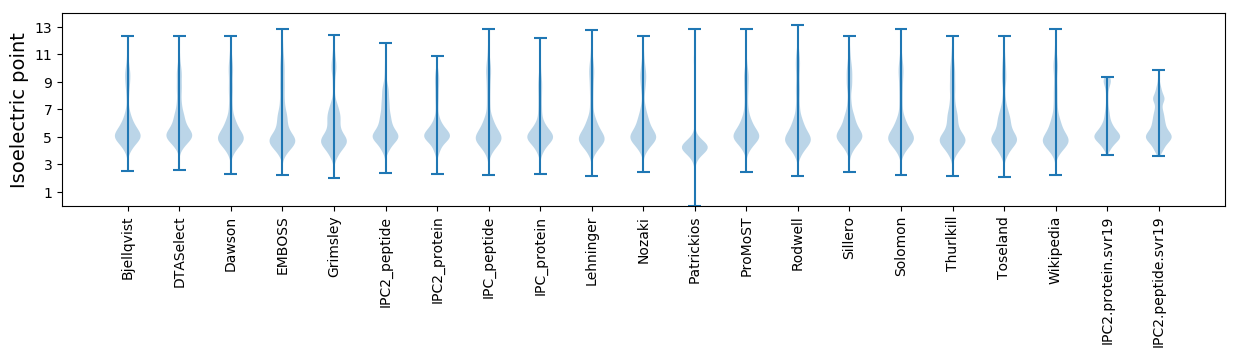
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C5ZTJ2|A0A5C5ZTJ2_9BACT Bifunctional purine biosynthesis protein PurH OS=Pseudobythopirellula maris OX=2527991 GN=purH PE=3 SV=1
MM1 pKa = 7.34LVDD4 pKa = 3.87TGMIDD9 pKa = 3.2TPRR12 pKa = 11.84TVRR15 pKa = 11.84LQSPSILTALAFTLLLAAPALGAKK39 pKa = 9.55YY40 pKa = 9.09PAYY43 pKa = 10.72VDD45 pKa = 3.71GSFTFGDD52 pKa = 3.93EE53 pKa = 4.09FAEE56 pKa = 4.31APYY59 pKa = 10.71GLEE62 pKa = 4.12STFSLSSNPNATKK75 pKa = 10.05TIYY78 pKa = 10.84LDD80 pKa = 3.28FTGFHH85 pKa = 6.44SVGNSWGHH93 pKa = 5.97NIVFPAYY100 pKa = 10.32DD101 pKa = 3.94LDD103 pKa = 4.8GDD105 pKa = 4.07SGSFSDD111 pKa = 4.66EE112 pKa = 3.77EE113 pKa = 4.76LIEE116 pKa = 4.12IQQIFQNVAEE126 pKa = 5.03DD127 pKa = 3.94YY128 pKa = 11.06IPFNVNVTTADD139 pKa = 3.44PGATALRR146 pKa = 11.84KK147 pKa = 10.21LGDD150 pKa = 4.09GDD152 pKa = 4.02THH154 pKa = 6.42WGMRR158 pKa = 11.84VVMTQATAGFGVGIGGDD175 pKa = 3.55SGSSRR180 pKa = 11.84FDD182 pKa = 3.55DD183 pKa = 4.76AADD186 pKa = 3.54NPVFTFNKK194 pKa = 9.04GARR197 pKa = 11.84KK198 pKa = 9.76GGLTASHH205 pKa = 6.24EE206 pKa = 4.41VGHH209 pKa = 5.96TFGLAHH215 pKa = 7.43DD216 pKa = 4.47SYY218 pKa = 11.73YY219 pKa = 10.87DD220 pKa = 3.53QEE222 pKa = 4.4YY223 pKa = 10.65HH224 pKa = 6.98PGTGGYY230 pKa = 10.41GRR232 pKa = 11.84TSWGPLMGAPFDD244 pKa = 4.36ASITQWSNGDD254 pKa = 3.5YY255 pKa = 10.67AGSSNTFDD263 pKa = 4.38DD264 pKa = 4.3YY265 pKa = 12.18SFITSKK271 pKa = 10.99GFGFRR276 pKa = 11.84EE277 pKa = 3.65DD278 pKa = 4.87DD279 pKa = 3.95YY280 pKa = 11.66LNSLADD286 pKa = 3.53PHH288 pKa = 8.11LLEE291 pKa = 6.09SIDD294 pKa = 4.15GEE296 pKa = 3.95ISEE299 pKa = 4.28WGIIEE304 pKa = 3.96EE305 pKa = 4.59RR306 pKa = 11.84YY307 pKa = 10.37DD308 pKa = 3.37RR309 pKa = 11.84DD310 pKa = 3.42YY311 pKa = 11.66FRR313 pKa = 11.84FTTTTGEE320 pKa = 3.86VSFDD324 pKa = 3.62VNGFAQDD331 pKa = 3.61PNIDD335 pKa = 3.92VGVVIYY341 pKa = 10.61DD342 pKa = 3.43SLGEE346 pKa = 4.21VVVEE350 pKa = 4.63LDD352 pKa = 3.74PSVTPNINYY361 pKa = 10.09DD362 pKa = 3.79LFLTAGDD369 pKa = 3.56YY370 pKa = 10.92TFFVDD375 pKa = 5.87GVDD378 pKa = 3.46NPGRR382 pKa = 11.84YY383 pKa = 8.89SDD385 pKa = 3.77YY386 pKa = 11.25GSLGFYY392 pKa = 10.08DD393 pKa = 3.77IQIGLPIAGDD403 pKa = 3.84LNSDD407 pKa = 3.9GFVDD411 pKa = 3.64TDD413 pKa = 3.15DD414 pKa = 3.79WLLLVAGAEE423 pKa = 4.24TDD425 pKa = 3.59LSGLSAEE432 pKa = 4.46EE433 pKa = 4.03AFLAGDD439 pKa = 4.03LNGDD443 pKa = 4.23GLNDD447 pKa = 3.36IYY449 pKa = 11.56DD450 pKa = 3.83FGLFKK455 pKa = 10.96DD456 pKa = 4.46AFTSAHH462 pKa = 5.89GAAEE466 pKa = 4.57FAALLAPEE474 pKa = 4.14PAGLGLATAMVLMLAGYY491 pKa = 7.33VRR493 pKa = 11.84CGVRR497 pKa = 11.84TNN499 pKa = 3.28
MM1 pKa = 7.34LVDD4 pKa = 3.87TGMIDD9 pKa = 3.2TPRR12 pKa = 11.84TVRR15 pKa = 11.84LQSPSILTALAFTLLLAAPALGAKK39 pKa = 9.55YY40 pKa = 9.09PAYY43 pKa = 10.72VDD45 pKa = 3.71GSFTFGDD52 pKa = 3.93EE53 pKa = 4.09FAEE56 pKa = 4.31APYY59 pKa = 10.71GLEE62 pKa = 4.12STFSLSSNPNATKK75 pKa = 10.05TIYY78 pKa = 10.84LDD80 pKa = 3.28FTGFHH85 pKa = 6.44SVGNSWGHH93 pKa = 5.97NIVFPAYY100 pKa = 10.32DD101 pKa = 3.94LDD103 pKa = 4.8GDD105 pKa = 4.07SGSFSDD111 pKa = 4.66EE112 pKa = 3.77EE113 pKa = 4.76LIEE116 pKa = 4.12IQQIFQNVAEE126 pKa = 5.03DD127 pKa = 3.94YY128 pKa = 11.06IPFNVNVTTADD139 pKa = 3.44PGATALRR146 pKa = 11.84KK147 pKa = 10.21LGDD150 pKa = 4.09GDD152 pKa = 4.02THH154 pKa = 6.42WGMRR158 pKa = 11.84VVMTQATAGFGVGIGGDD175 pKa = 3.55SGSSRR180 pKa = 11.84FDD182 pKa = 3.55DD183 pKa = 4.76AADD186 pKa = 3.54NPVFTFNKK194 pKa = 9.04GARR197 pKa = 11.84KK198 pKa = 9.76GGLTASHH205 pKa = 6.24EE206 pKa = 4.41VGHH209 pKa = 5.96TFGLAHH215 pKa = 7.43DD216 pKa = 4.47SYY218 pKa = 11.73YY219 pKa = 10.87DD220 pKa = 3.53QEE222 pKa = 4.4YY223 pKa = 10.65HH224 pKa = 6.98PGTGGYY230 pKa = 10.41GRR232 pKa = 11.84TSWGPLMGAPFDD244 pKa = 4.36ASITQWSNGDD254 pKa = 3.5YY255 pKa = 10.67AGSSNTFDD263 pKa = 4.38DD264 pKa = 4.3YY265 pKa = 12.18SFITSKK271 pKa = 10.99GFGFRR276 pKa = 11.84EE277 pKa = 3.65DD278 pKa = 4.87DD279 pKa = 3.95YY280 pKa = 11.66LNSLADD286 pKa = 3.53PHH288 pKa = 8.11LLEE291 pKa = 6.09SIDD294 pKa = 4.15GEE296 pKa = 3.95ISEE299 pKa = 4.28WGIIEE304 pKa = 3.96EE305 pKa = 4.59RR306 pKa = 11.84YY307 pKa = 10.37DD308 pKa = 3.37RR309 pKa = 11.84DD310 pKa = 3.42YY311 pKa = 11.66FRR313 pKa = 11.84FTTTTGEE320 pKa = 3.86VSFDD324 pKa = 3.62VNGFAQDD331 pKa = 3.61PNIDD335 pKa = 3.92VGVVIYY341 pKa = 10.61DD342 pKa = 3.43SLGEE346 pKa = 4.21VVVEE350 pKa = 4.63LDD352 pKa = 3.74PSVTPNINYY361 pKa = 10.09DD362 pKa = 3.79LFLTAGDD369 pKa = 3.56YY370 pKa = 10.92TFFVDD375 pKa = 5.87GVDD378 pKa = 3.46NPGRR382 pKa = 11.84YY383 pKa = 8.89SDD385 pKa = 3.77YY386 pKa = 11.25GSLGFYY392 pKa = 10.08DD393 pKa = 3.77IQIGLPIAGDD403 pKa = 3.84LNSDD407 pKa = 3.9GFVDD411 pKa = 3.64TDD413 pKa = 3.15DD414 pKa = 3.79WLLLVAGAEE423 pKa = 4.24TDD425 pKa = 3.59LSGLSAEE432 pKa = 4.46EE433 pKa = 4.03AFLAGDD439 pKa = 4.03LNGDD443 pKa = 4.23GLNDD447 pKa = 3.36IYY449 pKa = 11.56DD450 pKa = 3.83FGLFKK455 pKa = 10.96DD456 pKa = 4.46AFTSAHH462 pKa = 5.89GAAEE466 pKa = 4.57FAALLAPEE474 pKa = 4.14PAGLGLATAMVLMLAGYY491 pKa = 7.33VRR493 pKa = 11.84CGVRR497 pKa = 11.84TNN499 pKa = 3.28
Molecular weight: 53.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C5ZRC2|A0A5C5ZRC2_9BACT PSP1 C-terminal domain-containing protein OS=Pseudobythopirellula maris OX=2527991 GN=Mal64_04850 PE=4 SV=1
MM1 pKa = 7.8RR2 pKa = 11.84LANKK6 pKa = 9.79HH7 pKa = 5.74AMITGAASGIGRR19 pKa = 11.84AIAMRR24 pKa = 11.84LASRR28 pKa = 11.84MTHH31 pKa = 5.65LTLVDD36 pKa = 3.38IDD38 pKa = 3.77AAGLEE43 pKa = 4.31RR44 pKa = 11.84TARR47 pKa = 11.84EE48 pKa = 3.91ARR50 pKa = 11.84EE51 pKa = 3.78LGAAVDD57 pKa = 3.73SQRR60 pKa = 11.84RR61 pKa = 11.84DD62 pKa = 3.0LTLRR66 pKa = 11.84DD67 pKa = 3.64EE68 pKa = 4.58VDD70 pKa = 3.06ATVQEE75 pKa = 4.75SLDD78 pKa = 3.49QSAGVDD84 pKa = 3.52LLVNNAGVTYY94 pKa = 10.19HH95 pKa = 6.83GPTHH99 pKa = 7.05AMPAGDD105 pKa = 4.86LEE107 pKa = 4.8RR108 pKa = 11.84LMSLNLLAHH117 pKa = 5.88VRR119 pKa = 11.84LTHH122 pKa = 6.59GLLPSLLARR131 pKa = 11.84PEE133 pKa = 3.78AHH135 pKa = 6.31VLNVCSVLGLAGMPRR150 pKa = 11.84VSAYY154 pKa = 10.45CAGKK158 pKa = 9.16FAMVGFSEE166 pKa = 4.48SLRR169 pKa = 11.84SEE171 pKa = 4.17YY172 pKa = 10.96GRR174 pKa = 11.84VGLGVTAFCPGFVRR188 pKa = 11.84TEE190 pKa = 4.13LFGSAPLPEE199 pKa = 4.84GQSKK203 pKa = 10.11PKK205 pKa = 10.55SPPAWICLSPEE216 pKa = 3.57RR217 pKa = 11.84VARR220 pKa = 11.84RR221 pKa = 11.84AVLAVEE227 pKa = 4.55RR228 pKa = 11.84NRR230 pKa = 11.84GRR232 pKa = 11.84VVLDD236 pKa = 4.31PYY238 pKa = 10.4CWWAIGLKK246 pKa = 9.95RR247 pKa = 11.84RR248 pKa = 11.84LPAAFDD254 pKa = 3.43WALSLGRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84VEE266 pKa = 3.82RR267 pKa = 11.84KK268 pKa = 9.32RR269 pKa = 11.84RR270 pKa = 11.84EE271 pKa = 4.0LAALAPDD278 pKa = 4.0PQTALRR284 pKa = 11.84MKK286 pKa = 10.23LGLGEE291 pKa = 4.2RR292 pKa = 11.84PRR294 pKa = 11.84LARR297 pKa = 11.84AAA299 pKa = 3.71
MM1 pKa = 7.8RR2 pKa = 11.84LANKK6 pKa = 9.79HH7 pKa = 5.74AMITGAASGIGRR19 pKa = 11.84AIAMRR24 pKa = 11.84LASRR28 pKa = 11.84MTHH31 pKa = 5.65LTLVDD36 pKa = 3.38IDD38 pKa = 3.77AAGLEE43 pKa = 4.31RR44 pKa = 11.84TARR47 pKa = 11.84EE48 pKa = 3.91ARR50 pKa = 11.84EE51 pKa = 3.78LGAAVDD57 pKa = 3.73SQRR60 pKa = 11.84RR61 pKa = 11.84DD62 pKa = 3.0LTLRR66 pKa = 11.84DD67 pKa = 3.64EE68 pKa = 4.58VDD70 pKa = 3.06ATVQEE75 pKa = 4.75SLDD78 pKa = 3.49QSAGVDD84 pKa = 3.52LLVNNAGVTYY94 pKa = 10.19HH95 pKa = 6.83GPTHH99 pKa = 7.05AMPAGDD105 pKa = 4.86LEE107 pKa = 4.8RR108 pKa = 11.84LMSLNLLAHH117 pKa = 5.88VRR119 pKa = 11.84LTHH122 pKa = 6.59GLLPSLLARR131 pKa = 11.84PEE133 pKa = 3.78AHH135 pKa = 6.31VLNVCSVLGLAGMPRR150 pKa = 11.84VSAYY154 pKa = 10.45CAGKK158 pKa = 9.16FAMVGFSEE166 pKa = 4.48SLRR169 pKa = 11.84SEE171 pKa = 4.17YY172 pKa = 10.96GRR174 pKa = 11.84VGLGVTAFCPGFVRR188 pKa = 11.84TEE190 pKa = 4.13LFGSAPLPEE199 pKa = 4.84GQSKK203 pKa = 10.11PKK205 pKa = 10.55SPPAWICLSPEE216 pKa = 3.57RR217 pKa = 11.84VARR220 pKa = 11.84RR221 pKa = 11.84AVLAVEE227 pKa = 4.55RR228 pKa = 11.84NRR230 pKa = 11.84GRR232 pKa = 11.84VVLDD236 pKa = 4.31PYY238 pKa = 10.4CWWAIGLKK246 pKa = 9.95RR247 pKa = 11.84RR248 pKa = 11.84LPAAFDD254 pKa = 3.43WALSLGRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84VEE266 pKa = 3.82RR267 pKa = 11.84KK268 pKa = 9.32RR269 pKa = 11.84RR270 pKa = 11.84EE271 pKa = 4.0LAALAPDD278 pKa = 4.0PQTALRR284 pKa = 11.84MKK286 pKa = 10.23LGLGEE291 pKa = 4.2RR292 pKa = 11.84PRR294 pKa = 11.84LARR297 pKa = 11.84AAA299 pKa = 3.71
Molecular weight: 32.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1431816 |
30 |
7557 |
367.8 |
39.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.795 ± 0.054 | 1.051 ± 0.018 |
6.251 ± 0.04 | 6.715 ± 0.046 |
3.314 ± 0.023 | 8.764 ± 0.046 |
2.034 ± 0.015 | 3.885 ± 0.026 |
2.838 ± 0.032 | 10.041 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.03 ± 0.019 | 2.66 ± 0.03 |
5.507 ± 0.035 | 3.235 ± 0.024 |
7.04 ± 0.046 | 6.036 ± 0.033 |
5.372 ± 0.035 | 7.524 ± 0.031 |
1.493 ± 0.018 | 2.413 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
