
Wenzhou tombus-like virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
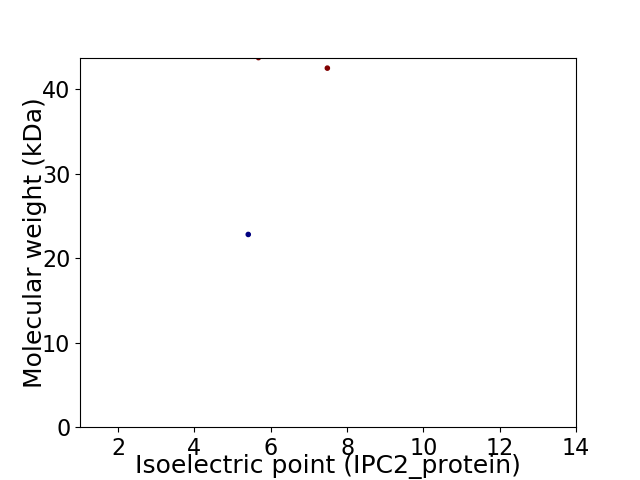
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
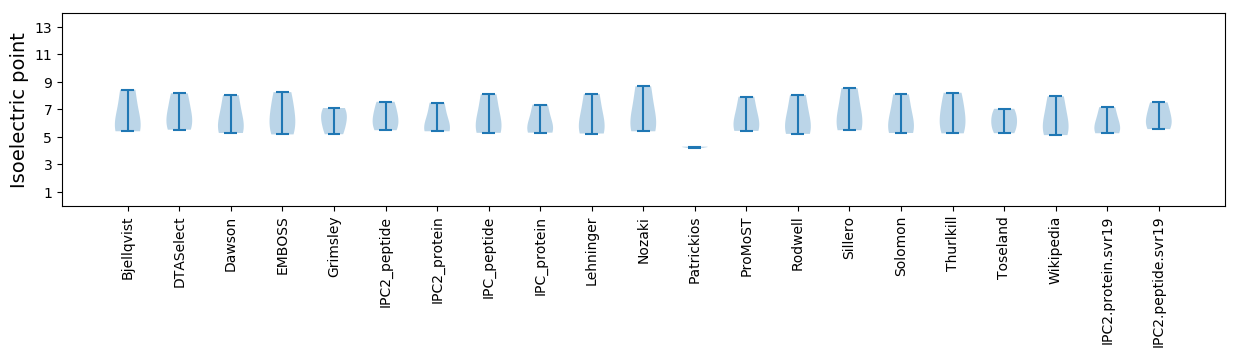
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH21|A0A1L3KH21_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 6 OX=1923676 PE=4 SV=1
MM1 pKa = 7.03KK2 pKa = 9.39TDD4 pKa = 3.55RR5 pKa = 11.84YY6 pKa = 10.49NFVVSYY12 pKa = 10.99GGYY15 pKa = 7.04TYY17 pKa = 11.42KK18 pKa = 10.65RR19 pKa = 11.84MEE21 pKa = 4.68FGTISVTLDD30 pKa = 3.08EE31 pKa = 5.04NEE33 pKa = 4.19VKK35 pKa = 10.64GVSRR39 pKa = 11.84PRR41 pKa = 11.84KK42 pKa = 6.04EE43 pKa = 4.22HH44 pKa = 6.16KK45 pKa = 10.31SISAAHH51 pKa = 5.99TLYY54 pKa = 10.24PDD56 pKa = 3.57YY57 pKa = 11.15EE58 pKa = 4.46SLLNASQEE66 pKa = 4.08QTLSEE71 pKa = 3.96EE72 pKa = 3.9SDD74 pKa = 3.48AIKK77 pKa = 10.35AYY79 pKa = 9.44EE80 pKa = 3.85QAANRR85 pKa = 11.84QGDD88 pKa = 3.87DD89 pKa = 3.72SEE91 pKa = 5.7SDD93 pKa = 3.89AEE95 pKa = 4.32VLPPPGYY102 pKa = 10.24EE103 pKa = 3.79PEE105 pKa = 4.59TIVGLPPPGPGRR117 pKa = 11.84VPYY120 pKa = 10.16RR121 pKa = 11.84ADD123 pKa = 4.48PISIPNVVVKK133 pKa = 9.69PSNNRR138 pKa = 11.84NTGSFLSRR146 pKa = 11.84LVEE149 pKa = 4.25SQKK152 pKa = 9.34TAPRR156 pKa = 11.84DD157 pKa = 3.6EE158 pKa = 4.45EE159 pKa = 4.44TLSVTSARR167 pKa = 11.84TTGSLTPEE175 pKa = 3.52EE176 pKa = 4.83RR177 pKa = 11.84IEE179 pKa = 3.64YY180 pKa = 8.0WRR182 pKa = 11.84IYY184 pKa = 10.01RR185 pKa = 11.84SRR187 pKa = 11.84GKK189 pKa = 10.23AAADD193 pKa = 4.38FYY195 pKa = 11.41KK196 pKa = 10.59KK197 pKa = 10.35RR198 pKa = 11.84IEE200 pKa = 4.56DD201 pKa = 3.53NPGG204 pKa = 2.74
MM1 pKa = 7.03KK2 pKa = 9.39TDD4 pKa = 3.55RR5 pKa = 11.84YY6 pKa = 10.49NFVVSYY12 pKa = 10.99GGYY15 pKa = 7.04TYY17 pKa = 11.42KK18 pKa = 10.65RR19 pKa = 11.84MEE21 pKa = 4.68FGTISVTLDD30 pKa = 3.08EE31 pKa = 5.04NEE33 pKa = 4.19VKK35 pKa = 10.64GVSRR39 pKa = 11.84PRR41 pKa = 11.84KK42 pKa = 6.04EE43 pKa = 4.22HH44 pKa = 6.16KK45 pKa = 10.31SISAAHH51 pKa = 5.99TLYY54 pKa = 10.24PDD56 pKa = 3.57YY57 pKa = 11.15EE58 pKa = 4.46SLLNASQEE66 pKa = 4.08QTLSEE71 pKa = 3.96EE72 pKa = 3.9SDD74 pKa = 3.48AIKK77 pKa = 10.35AYY79 pKa = 9.44EE80 pKa = 3.85QAANRR85 pKa = 11.84QGDD88 pKa = 3.87DD89 pKa = 3.72SEE91 pKa = 5.7SDD93 pKa = 3.89AEE95 pKa = 4.32VLPPPGYY102 pKa = 10.24EE103 pKa = 3.79PEE105 pKa = 4.59TIVGLPPPGPGRR117 pKa = 11.84VPYY120 pKa = 10.16RR121 pKa = 11.84ADD123 pKa = 4.48PISIPNVVVKK133 pKa = 9.69PSNNRR138 pKa = 11.84NTGSFLSRR146 pKa = 11.84LVEE149 pKa = 4.25SQKK152 pKa = 9.34TAPRR156 pKa = 11.84DD157 pKa = 3.6EE158 pKa = 4.45EE159 pKa = 4.44TLSVTSARR167 pKa = 11.84TTGSLTPEE175 pKa = 3.52EE176 pKa = 4.83RR177 pKa = 11.84IEE179 pKa = 3.64YY180 pKa = 8.0WRR182 pKa = 11.84IYY184 pKa = 10.01RR185 pKa = 11.84SRR187 pKa = 11.84GKK189 pKa = 10.23AAADD193 pKa = 4.38FYY195 pKa = 11.41KK196 pKa = 10.59KK197 pKa = 10.35RR198 pKa = 11.84IEE200 pKa = 4.56DD201 pKa = 3.53NPGG204 pKa = 2.74
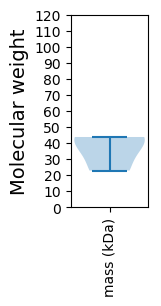
Molecular weight: 22.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH21|A0A1L3KH21_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 6 OX=1923676 PE=4 SV=1
MM1 pKa = 7.74IKK3 pKa = 10.27PIAPRR8 pKa = 11.84LICPRR13 pKa = 11.84TKK15 pKa = 10.14RR16 pKa = 11.84YY17 pKa = 9.81NVILGTRR24 pKa = 11.84LKK26 pKa = 10.63FNEE29 pKa = 3.93KK30 pKa = 10.03RR31 pKa = 11.84IMRR34 pKa = 11.84AIDD37 pKa = 3.54SVFGSPTVLSSYY49 pKa = 11.4DD50 pKa = 3.13SFAQGRR56 pKa = 11.84IIARR60 pKa = 11.84KK61 pKa = 7.61WAMFNNPVAIGVDD74 pKa = 3.46ASRR77 pKa = 11.84FDD79 pKa = 3.39QHH81 pKa = 8.72VSVQALQWEE90 pKa = 4.31HH91 pKa = 6.95SIYY94 pKa = 10.81NAIFHH99 pKa = 6.91DD100 pKa = 4.18PEE102 pKa = 4.16LRR104 pKa = 11.84EE105 pKa = 4.25ALSHH109 pKa = 5.22QLKK112 pKa = 10.8NNISLFVEE120 pKa = 4.35DD121 pKa = 3.82KK122 pKa = 10.25MIRR125 pKa = 11.84FKK127 pKa = 11.44VDD129 pKa = 2.51GHH131 pKa = 6.48RR132 pKa = 11.84MSGDD136 pKa = 2.98INTSMGNKK144 pKa = 10.23LIMCGLMHH152 pKa = 7.29HH153 pKa = 6.02YY154 pKa = 10.19FKK156 pKa = 10.71TLGVRR161 pKa = 11.84AEE163 pKa = 4.19LCNNGDD169 pKa = 3.8DD170 pKa = 4.92CVIICEE176 pKa = 4.47NKK178 pKa = 10.21DD179 pKa = 2.86KK180 pKa = 11.28HH181 pKa = 7.26LFDD184 pKa = 5.43NIYY187 pKa = 10.7EE188 pKa = 4.33HH189 pKa = 7.07FLQFGFNMVTEE200 pKa = 4.3PVVEE204 pKa = 4.15CLEE207 pKa = 4.11KK208 pKa = 11.32LEE210 pKa = 4.64FCQSRR215 pKa = 11.84PICVNGAWRR224 pKa = 11.84MVRR227 pKa = 11.84RR228 pKa = 11.84PDD230 pKa = 4.29SIAKK234 pKa = 9.99DD235 pKa = 3.52SHH237 pKa = 6.31TLLSMLNAEE246 pKa = 4.5DD247 pKa = 3.79VKK249 pKa = 11.57SFMSATAQCGLVLNSGVPILEE270 pKa = 3.83AFYY273 pKa = 10.33RR274 pKa = 11.84CLYY277 pKa = 10.12RR278 pKa = 11.84SSGYY282 pKa = 10.69KK283 pKa = 9.91KK284 pKa = 9.59VTEE287 pKa = 4.4EE288 pKa = 4.06YY289 pKa = 8.37IKK291 pKa = 11.05AVIGYY296 pKa = 9.48GNEE299 pKa = 3.64EE300 pKa = 4.03KK301 pKa = 10.93LQGRR305 pKa = 11.84RR306 pKa = 11.84NPVSIPVTDD315 pKa = 4.01STRR318 pKa = 11.84MSYY321 pKa = 9.33WEE323 pKa = 4.34SFGVDD328 pKa = 3.65PKK330 pKa = 9.66TQQIIEE336 pKa = 4.42RR337 pKa = 11.84YY338 pKa = 8.92YY339 pKa = 11.45DD340 pKa = 3.97DD341 pKa = 4.54LTVSTQLQPVKK352 pKa = 10.08LTTPHH357 pKa = 6.1LQSILLEE364 pKa = 4.49IPQHH368 pKa = 4.73PTII371 pKa = 4.71
MM1 pKa = 7.74IKK3 pKa = 10.27PIAPRR8 pKa = 11.84LICPRR13 pKa = 11.84TKK15 pKa = 10.14RR16 pKa = 11.84YY17 pKa = 9.81NVILGTRR24 pKa = 11.84LKK26 pKa = 10.63FNEE29 pKa = 3.93KK30 pKa = 10.03RR31 pKa = 11.84IMRR34 pKa = 11.84AIDD37 pKa = 3.54SVFGSPTVLSSYY49 pKa = 11.4DD50 pKa = 3.13SFAQGRR56 pKa = 11.84IIARR60 pKa = 11.84KK61 pKa = 7.61WAMFNNPVAIGVDD74 pKa = 3.46ASRR77 pKa = 11.84FDD79 pKa = 3.39QHH81 pKa = 8.72VSVQALQWEE90 pKa = 4.31HH91 pKa = 6.95SIYY94 pKa = 10.81NAIFHH99 pKa = 6.91DD100 pKa = 4.18PEE102 pKa = 4.16LRR104 pKa = 11.84EE105 pKa = 4.25ALSHH109 pKa = 5.22QLKK112 pKa = 10.8NNISLFVEE120 pKa = 4.35DD121 pKa = 3.82KK122 pKa = 10.25MIRR125 pKa = 11.84FKK127 pKa = 11.44VDD129 pKa = 2.51GHH131 pKa = 6.48RR132 pKa = 11.84MSGDD136 pKa = 2.98INTSMGNKK144 pKa = 10.23LIMCGLMHH152 pKa = 7.29HH153 pKa = 6.02YY154 pKa = 10.19FKK156 pKa = 10.71TLGVRR161 pKa = 11.84AEE163 pKa = 4.19LCNNGDD169 pKa = 3.8DD170 pKa = 4.92CVIICEE176 pKa = 4.47NKK178 pKa = 10.21DD179 pKa = 2.86KK180 pKa = 11.28HH181 pKa = 7.26LFDD184 pKa = 5.43NIYY187 pKa = 10.7EE188 pKa = 4.33HH189 pKa = 7.07FLQFGFNMVTEE200 pKa = 4.3PVVEE204 pKa = 4.15CLEE207 pKa = 4.11KK208 pKa = 11.32LEE210 pKa = 4.64FCQSRR215 pKa = 11.84PICVNGAWRR224 pKa = 11.84MVRR227 pKa = 11.84RR228 pKa = 11.84PDD230 pKa = 4.29SIAKK234 pKa = 9.99DD235 pKa = 3.52SHH237 pKa = 6.31TLLSMLNAEE246 pKa = 4.5DD247 pKa = 3.79VKK249 pKa = 11.57SFMSATAQCGLVLNSGVPILEE270 pKa = 3.83AFYY273 pKa = 10.33RR274 pKa = 11.84CLYY277 pKa = 10.12RR278 pKa = 11.84SSGYY282 pKa = 10.69KK283 pKa = 9.91KK284 pKa = 9.59VTEE287 pKa = 4.4EE288 pKa = 4.06YY289 pKa = 8.37IKK291 pKa = 11.05AVIGYY296 pKa = 9.48GNEE299 pKa = 3.64EE300 pKa = 4.03KK301 pKa = 10.93LQGRR305 pKa = 11.84RR306 pKa = 11.84NPVSIPVTDD315 pKa = 4.01STRR318 pKa = 11.84MSYY321 pKa = 9.33WEE323 pKa = 4.34SFGVDD328 pKa = 3.65PKK330 pKa = 9.66TQQIIEE336 pKa = 4.42RR337 pKa = 11.84YY338 pKa = 8.92YY339 pKa = 11.45DD340 pKa = 3.97DD341 pKa = 4.54LTVSTQLQPVKK352 pKa = 10.08LTTPHH357 pKa = 6.1LQSILLEE364 pKa = 4.49IPQHH368 pKa = 4.73PTII371 pKa = 4.71
Molecular weight: 42.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
954 |
204 |
379 |
318.0 |
36.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.128 ± 1.393 | 1.363 ± 0.717 |
5.031 ± 0.152 | 8.386 ± 1.471 |
3.354 ± 0.668 | 4.612 ± 0.771 |
2.83 ± 0.555 | 5.975 ± 0.981 |
6.184 ± 0.49 | 8.281 ± 0.946 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.935 ± 0.587 | 4.088 ± 0.669 |
5.136 ± 0.968 | 3.774 ± 0.398 |
6.394 ± 0.445 | 7.338 ± 0.809 |
5.346 ± 0.508 | 6.394 ± 0.361 |
1.258 ± 0.319 | 4.193 ± 0.53 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
