
CRESS virus sp. ctfsu28
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; unclassified Cressdnaviricota; CRESS viruses
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
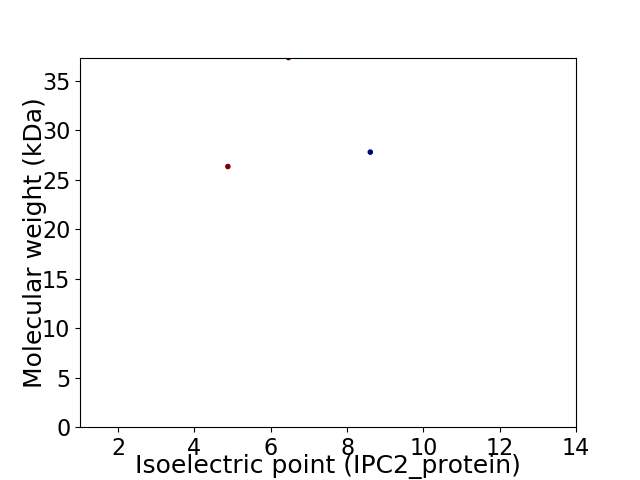
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
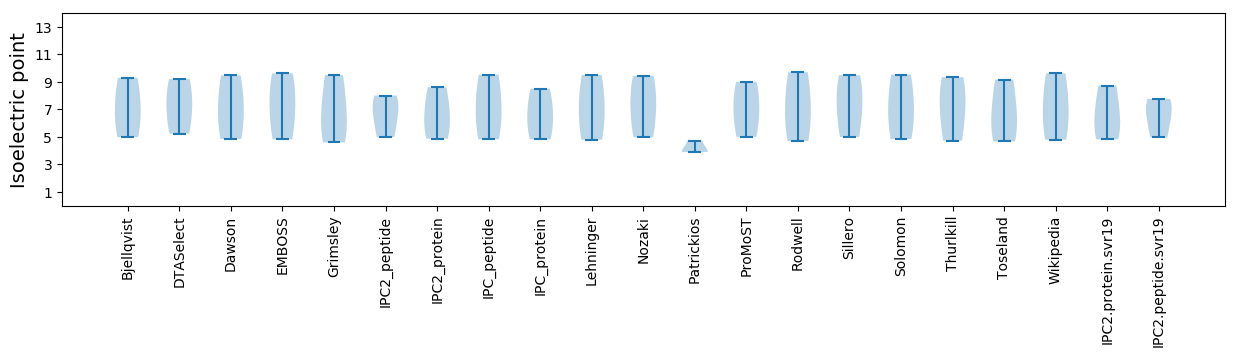
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W9N6|A0A5Q2W9N6_9VIRU ATP-dependent helicase Rep OS=CRESS virus sp. ctfsu28 OX=2656685 PE=3 SV=1
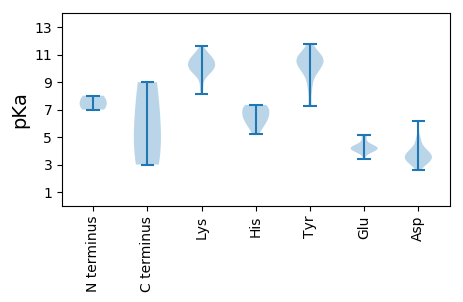
MM1 pKa = 7.0AHH3 pKa = 7.3KK4 pKa = 10.23RR5 pKa = 11.84KK6 pKa = 9.16QWFYY10 pKa = 10.19GSNSVRR16 pKa = 11.84RR17 pKa = 11.84SVRR20 pKa = 11.84QQEE23 pKa = 4.33DD24 pKa = 4.24GNTPEE29 pKa = 5.17LKK31 pKa = 10.08MYY33 pKa = 9.7DD34 pKa = 3.59QSVLDD39 pKa = 3.95TLIGTGTSWSGADD52 pKa = 4.01MDD54 pKa = 4.97PSSNTLFSPNLGVDD68 pKa = 3.41MGQRR72 pKa = 11.84IGRR75 pKa = 11.84RR76 pKa = 11.84CAVVGLDD83 pKa = 3.16MNGFIDD89 pKa = 5.79CNDD92 pKa = 2.98QVLAVGADD100 pKa = 3.47PAAICRR106 pKa = 11.84IIVFQDD112 pKa = 3.26TQTKK116 pKa = 9.83GVQALGNLVMEE127 pKa = 5.12AQTADD132 pKa = 3.36AFHH135 pKa = 7.22VITAFPSRR143 pKa = 11.84DD144 pKa = 3.36AIGRR148 pKa = 11.84FIILKK153 pKa = 10.4DD154 pKa = 3.4MVYY157 pKa = 11.17VMQNPAFSSLGSSGVTYY174 pKa = 9.98DD175 pKa = 3.25QQGIAKK181 pKa = 8.49WFSFRR186 pKa = 11.84IEE188 pKa = 3.7FDD190 pKa = 3.27EE191 pKa = 4.08PVIVHH196 pKa = 6.47FDD198 pKa = 3.21ASDD201 pKa = 3.24AGTVADD207 pKa = 3.75VLDD210 pKa = 3.97NSFHH214 pKa = 7.34VMASNSSSGLSPRR227 pKa = 11.84LTYY230 pKa = 7.32TTRR233 pKa = 11.84VYY235 pKa = 11.05YY236 pKa = 10.81YY237 pKa = 10.8DD238 pKa = 3.51FF239 pKa = 5.41
MM1 pKa = 7.0AHH3 pKa = 7.3KK4 pKa = 10.23RR5 pKa = 11.84KK6 pKa = 9.16QWFYY10 pKa = 10.19GSNSVRR16 pKa = 11.84RR17 pKa = 11.84SVRR20 pKa = 11.84QQEE23 pKa = 4.33DD24 pKa = 4.24GNTPEE29 pKa = 5.17LKK31 pKa = 10.08MYY33 pKa = 9.7DD34 pKa = 3.59QSVLDD39 pKa = 3.95TLIGTGTSWSGADD52 pKa = 4.01MDD54 pKa = 4.97PSSNTLFSPNLGVDD68 pKa = 3.41MGQRR72 pKa = 11.84IGRR75 pKa = 11.84RR76 pKa = 11.84CAVVGLDD83 pKa = 3.16MNGFIDD89 pKa = 5.79CNDD92 pKa = 2.98QVLAVGADD100 pKa = 3.47PAAICRR106 pKa = 11.84IIVFQDD112 pKa = 3.26TQTKK116 pKa = 9.83GVQALGNLVMEE127 pKa = 5.12AQTADD132 pKa = 3.36AFHH135 pKa = 7.22VITAFPSRR143 pKa = 11.84DD144 pKa = 3.36AIGRR148 pKa = 11.84FIILKK153 pKa = 10.4DD154 pKa = 3.4MVYY157 pKa = 11.17VMQNPAFSSLGSSGVTYY174 pKa = 9.98DD175 pKa = 3.25QQGIAKK181 pKa = 8.49WFSFRR186 pKa = 11.84IEE188 pKa = 3.7FDD190 pKa = 3.27EE191 pKa = 4.08PVIVHH196 pKa = 6.47FDD198 pKa = 3.21ASDD201 pKa = 3.24AGTVADD207 pKa = 3.75VLDD210 pKa = 3.97NSFHH214 pKa = 7.34VMASNSSSGLSPRR227 pKa = 11.84LTYY230 pKa = 7.32TTRR233 pKa = 11.84VYY235 pKa = 11.05YY236 pKa = 10.81YY237 pKa = 10.8DD238 pKa = 3.51FF239 pKa = 5.41
Molecular weight: 26.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2WAC3|A0A5Q2WAC3_9VIRU Capsid protein OS=CRESS virus sp. ctfsu28 OX=2656685 PE=4 SV=1
MM1 pKa = 7.48AGKK4 pKa = 10.01RR5 pKa = 11.84KK6 pKa = 10.11YY7 pKa = 10.03KK8 pKa = 10.48ADD10 pKa = 3.72YY11 pKa = 9.73KK12 pKa = 11.09KK13 pKa = 10.91DD14 pKa = 3.2GAGVKK19 pKa = 9.62KK20 pKa = 10.73YY21 pKa = 10.16KK22 pKa = 10.76GSASQQNYY30 pKa = 8.3IKK32 pKa = 10.48RR33 pKa = 11.84EE34 pKa = 4.02YY35 pKa = 10.47VPRR38 pKa = 11.84TPGGAVVSEE47 pKa = 4.38LKK49 pKa = 11.03VIDD52 pKa = 3.97SFLANTVLSAPTSWANTEE70 pKa = 3.93VDD72 pKa = 4.09PATLNCLFAPGPGTGLNQRR91 pKa = 11.84IGRR94 pKa = 11.84KK95 pKa = 9.08VAVKK99 pKa = 9.65KK100 pKa = 10.78LKK102 pKa = 10.19VRR104 pKa = 11.84GLITCASQVDD114 pKa = 4.04QTTPDD119 pKa = 3.05NGCQVRR125 pKa = 11.84LIFYY129 pKa = 10.65QDD131 pKa = 3.16MQTNGAQAQGEE142 pKa = 4.54DD143 pKa = 4.87LMDD146 pKa = 3.73TPTASVQGVFNSFQNTASFGRR167 pKa = 11.84FRR169 pKa = 11.84VLKK172 pKa = 10.4DD173 pKa = 2.89MCIEE177 pKa = 3.94INNPNISWDD186 pKa = 3.69GTNLEE191 pKa = 3.87QQGIVKK197 pKa = 10.11RR198 pKa = 11.84FKK200 pKa = 9.6FSKK203 pKa = 9.76VFKK206 pKa = 9.63EE207 pKa = 4.04PVVVHH212 pKa = 5.83YY213 pKa = 10.25NATGGTTVADD223 pKa = 3.62VVDD226 pKa = 4.15NSFHH230 pKa = 7.11AIAVVNSAALVPNLSYY246 pKa = 10.94VCRR249 pKa = 11.84VTYY252 pKa = 10.55TDD254 pKa = 3.1VV255 pKa = 3.0
MM1 pKa = 7.48AGKK4 pKa = 10.01RR5 pKa = 11.84KK6 pKa = 10.11YY7 pKa = 10.03KK8 pKa = 10.48ADD10 pKa = 3.72YY11 pKa = 9.73KK12 pKa = 11.09KK13 pKa = 10.91DD14 pKa = 3.2GAGVKK19 pKa = 9.62KK20 pKa = 10.73YY21 pKa = 10.16KK22 pKa = 10.76GSASQQNYY30 pKa = 8.3IKK32 pKa = 10.48RR33 pKa = 11.84EE34 pKa = 4.02YY35 pKa = 10.47VPRR38 pKa = 11.84TPGGAVVSEE47 pKa = 4.38LKK49 pKa = 11.03VIDD52 pKa = 3.97SFLANTVLSAPTSWANTEE70 pKa = 3.93VDD72 pKa = 4.09PATLNCLFAPGPGTGLNQRR91 pKa = 11.84IGRR94 pKa = 11.84KK95 pKa = 9.08VAVKK99 pKa = 9.65KK100 pKa = 10.78LKK102 pKa = 10.19VRR104 pKa = 11.84GLITCASQVDD114 pKa = 4.04QTTPDD119 pKa = 3.05NGCQVRR125 pKa = 11.84LIFYY129 pKa = 10.65QDD131 pKa = 3.16MQTNGAQAQGEE142 pKa = 4.54DD143 pKa = 4.87LMDD146 pKa = 3.73TPTASVQGVFNSFQNTASFGRR167 pKa = 11.84FRR169 pKa = 11.84VLKK172 pKa = 10.4DD173 pKa = 2.89MCIEE177 pKa = 3.94INNPNISWDD186 pKa = 3.69GTNLEE191 pKa = 3.87QQGIVKK197 pKa = 10.11RR198 pKa = 11.84FKK200 pKa = 9.6FSKK203 pKa = 9.76VFKK206 pKa = 9.63EE207 pKa = 4.04PVVVHH212 pKa = 5.83YY213 pKa = 10.25NATGGTTVADD223 pKa = 3.62VVDD226 pKa = 4.15NSFHH230 pKa = 7.11AIAVVNSAALVPNLSYY246 pKa = 10.94VCRR249 pKa = 11.84VTYY252 pKa = 10.55TDD254 pKa = 3.1VV255 pKa = 3.0
Molecular weight: 27.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
816 |
239 |
322 |
272.0 |
30.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.985 ± 0.962 | 2.328 ± 0.538 |
7.23 ± 0.742 | 4.412 ± 1.444 |
4.044 ± 0.816 | 8.088 ± 0.142 |
1.593 ± 0.336 | 5.025 ± 0.429 |
6.618 ± 1.585 | 5.882 ± 0.175 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.574 ± 0.492 | 5.392 ± 0.556 |
3.676 ± 0.247 | 3.676 ± 1.295 |
5.27 ± 0.387 | 6.373 ± 1.103 |
5.515 ± 0.912 | 8.701 ± 1.173 |
1.961 ± 0.693 | 4.657 ± 0.881 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
