
Hubei diptera virus 15
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
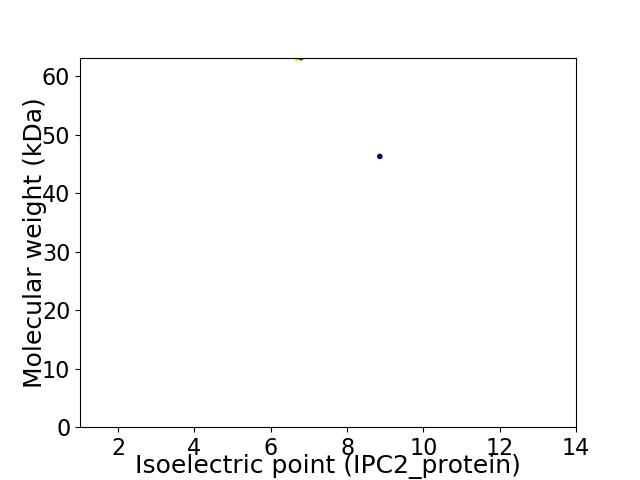
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGG9|A0A1L3KGG9_9VIRU RNA-directed RNA polymerase OS=Hubei diptera virus 15 OX=1922876 PE=4 SV=1
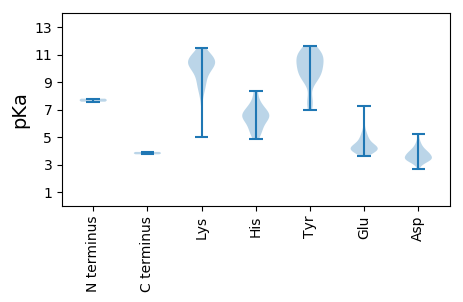
MM1 pKa = 7.79NIRR4 pKa = 11.84EE5 pKa = 4.22VYY7 pKa = 10.39NYY9 pKa = 10.85LFGEE13 pKa = 5.42GSAQDD18 pKa = 3.41QTEE21 pKa = 4.08DD22 pKa = 3.39EE23 pKa = 4.23LRR25 pKa = 11.84KK26 pKa = 10.15QIEE29 pKa = 4.32SLAKK33 pKa = 10.49VLEE36 pKa = 4.08QSHH39 pKa = 6.61KK40 pKa = 10.37IKK42 pKa = 10.73EE43 pKa = 4.16LMAQQPQPGDD53 pKa = 3.38TPTKK57 pKa = 9.85TKK59 pKa = 11.1EE60 pKa = 3.64EE61 pKa = 4.2LNYY64 pKa = 10.03VRR66 pKa = 11.84AKK68 pKa = 9.61ATAEE72 pKa = 4.15TLEE75 pKa = 4.29HH76 pKa = 6.62LAVIDD81 pKa = 5.24DD82 pKa = 3.91KK83 pKa = 11.09TLPPSVEE90 pKa = 4.36DD91 pKa = 3.84GLAFLKK97 pKa = 9.52TRR99 pKa = 11.84CEE101 pKa = 4.19GKK103 pKa = 10.45KK104 pKa = 10.05VDD106 pKa = 3.59HH107 pKa = 6.99TYY109 pKa = 11.05NAYY112 pKa = 9.65CASIVRR118 pKa = 11.84QYY120 pKa = 11.42LQTRR124 pKa = 11.84KK125 pKa = 8.5KK126 pKa = 8.74TSNKK130 pKa = 8.74MEE132 pKa = 4.11YY133 pKa = 10.53AIMNSIMNIHH143 pKa = 7.52LINLRR148 pKa = 11.84PQYY151 pKa = 10.94DD152 pKa = 3.78YY153 pKa = 11.65DD154 pKa = 3.29LHH156 pKa = 8.32EE157 pKa = 4.26IMKK160 pKa = 10.92YY161 pKa = 9.99NDD163 pKa = 4.22CINYY167 pKa = 7.61VQRR170 pKa = 11.84KK171 pKa = 5.02TFRR174 pKa = 11.84LFNSIYY180 pKa = 9.67EE181 pKa = 4.17IPFTHH186 pKa = 7.29INLEE190 pKa = 4.13PLNITALSLWNALPKK205 pKa = 10.15PLSKK209 pKa = 10.39FLQVLLLIITTYY221 pKa = 10.41FVVSGIILPSIIHH234 pKa = 6.33LTTSTTHH241 pKa = 5.6SQPPPPLQPMPIVTDD256 pKa = 3.3QSGWMSTTAHH266 pKa = 6.52YY267 pKa = 7.66LTTTACYY274 pKa = 9.84AGSSVAYY281 pKa = 9.9SLGAMTSTLSRR292 pKa = 11.84WIAPASASSTWIPSWIWKK310 pKa = 9.3PEE312 pKa = 3.91EE313 pKa = 3.98PSLIEE318 pKa = 4.12SLHH321 pKa = 5.49SQSSKK326 pKa = 9.36RR327 pKa = 11.84WEE329 pKa = 4.07QISTKK334 pKa = 10.6LHH336 pKa = 6.73DD337 pKa = 4.79LFKK340 pKa = 10.88LATPPLTWLLDD351 pKa = 3.53VTSNHH356 pKa = 6.06LNALSSSAIISAKK369 pKa = 10.55EE370 pKa = 3.92LMTKK374 pKa = 10.37LAPKK378 pKa = 8.51WHH380 pKa = 6.82ASVEE384 pKa = 4.68SINTTLRR391 pKa = 11.84EE392 pKa = 4.07TILPSMHH399 pKa = 5.96TSLWKK404 pKa = 10.63CYY406 pKa = 10.52DD407 pKa = 3.4YY408 pKa = 10.99ATDD411 pKa = 4.62FIYY414 pKa = 10.39GASTTTASYY423 pKa = 10.17YY424 pKa = 10.89DD425 pKa = 4.18SVVKK429 pKa = 9.95PYY431 pKa = 9.64ATEE434 pKa = 4.96LYY436 pKa = 10.17HH437 pKa = 8.26GMVNATPSMARR448 pKa = 11.84ACQVTLTRR456 pKa = 11.84VLVTVLSITTSLGLFSLTCISVAMQLSMEE485 pKa = 4.84TITSFLQTYY494 pKa = 9.25QLMAVQQQPHH504 pKa = 6.38SDD506 pKa = 3.14VTTWIQSFFHH516 pKa = 5.43QVVIFTQLAFVVLSYY531 pKa = 8.81FTTRR535 pKa = 11.84TVIQRR540 pKa = 11.84WDD542 pKa = 4.4LIPYY546 pKa = 9.31DD547 pKa = 3.81SRR549 pKa = 11.84RR550 pKa = 11.84SMVALTT556 pKa = 3.92
MM1 pKa = 7.79NIRR4 pKa = 11.84EE5 pKa = 4.22VYY7 pKa = 10.39NYY9 pKa = 10.85LFGEE13 pKa = 5.42GSAQDD18 pKa = 3.41QTEE21 pKa = 4.08DD22 pKa = 3.39EE23 pKa = 4.23LRR25 pKa = 11.84KK26 pKa = 10.15QIEE29 pKa = 4.32SLAKK33 pKa = 10.49VLEE36 pKa = 4.08QSHH39 pKa = 6.61KK40 pKa = 10.37IKK42 pKa = 10.73EE43 pKa = 4.16LMAQQPQPGDD53 pKa = 3.38TPTKK57 pKa = 9.85TKK59 pKa = 11.1EE60 pKa = 3.64EE61 pKa = 4.2LNYY64 pKa = 10.03VRR66 pKa = 11.84AKK68 pKa = 9.61ATAEE72 pKa = 4.15TLEE75 pKa = 4.29HH76 pKa = 6.62LAVIDD81 pKa = 5.24DD82 pKa = 3.91KK83 pKa = 11.09TLPPSVEE90 pKa = 4.36DD91 pKa = 3.84GLAFLKK97 pKa = 9.52TRR99 pKa = 11.84CEE101 pKa = 4.19GKK103 pKa = 10.45KK104 pKa = 10.05VDD106 pKa = 3.59HH107 pKa = 6.99TYY109 pKa = 11.05NAYY112 pKa = 9.65CASIVRR118 pKa = 11.84QYY120 pKa = 11.42LQTRR124 pKa = 11.84KK125 pKa = 8.5KK126 pKa = 8.74TSNKK130 pKa = 8.74MEE132 pKa = 4.11YY133 pKa = 10.53AIMNSIMNIHH143 pKa = 7.52LINLRR148 pKa = 11.84PQYY151 pKa = 10.94DD152 pKa = 3.78YY153 pKa = 11.65DD154 pKa = 3.29LHH156 pKa = 8.32EE157 pKa = 4.26IMKK160 pKa = 10.92YY161 pKa = 9.99NDD163 pKa = 4.22CINYY167 pKa = 7.61VQRR170 pKa = 11.84KK171 pKa = 5.02TFRR174 pKa = 11.84LFNSIYY180 pKa = 9.67EE181 pKa = 4.17IPFTHH186 pKa = 7.29INLEE190 pKa = 4.13PLNITALSLWNALPKK205 pKa = 10.15PLSKK209 pKa = 10.39FLQVLLLIITTYY221 pKa = 10.41FVVSGIILPSIIHH234 pKa = 6.33LTTSTTHH241 pKa = 5.6SQPPPPLQPMPIVTDD256 pKa = 3.3QSGWMSTTAHH266 pKa = 6.52YY267 pKa = 7.66LTTTACYY274 pKa = 9.84AGSSVAYY281 pKa = 9.9SLGAMTSTLSRR292 pKa = 11.84WIAPASASSTWIPSWIWKK310 pKa = 9.3PEE312 pKa = 3.91EE313 pKa = 3.98PSLIEE318 pKa = 4.12SLHH321 pKa = 5.49SQSSKK326 pKa = 9.36RR327 pKa = 11.84WEE329 pKa = 4.07QISTKK334 pKa = 10.6LHH336 pKa = 6.73DD337 pKa = 4.79LFKK340 pKa = 10.88LATPPLTWLLDD351 pKa = 3.53VTSNHH356 pKa = 6.06LNALSSSAIISAKK369 pKa = 10.55EE370 pKa = 3.92LMTKK374 pKa = 10.37LAPKK378 pKa = 8.51WHH380 pKa = 6.82ASVEE384 pKa = 4.68SINTTLRR391 pKa = 11.84EE392 pKa = 4.07TILPSMHH399 pKa = 5.96TSLWKK404 pKa = 10.63CYY406 pKa = 10.52DD407 pKa = 3.4YY408 pKa = 10.99ATDD411 pKa = 4.62FIYY414 pKa = 10.39GASTTTASYY423 pKa = 10.17YY424 pKa = 10.89DD425 pKa = 4.18SVVKK429 pKa = 9.95PYY431 pKa = 9.64ATEE434 pKa = 4.96LYY436 pKa = 10.17HH437 pKa = 8.26GMVNATPSMARR448 pKa = 11.84ACQVTLTRR456 pKa = 11.84VLVTVLSITTSLGLFSLTCISVAMQLSMEE485 pKa = 4.84TITSFLQTYY494 pKa = 9.25QLMAVQQQPHH504 pKa = 6.38SDD506 pKa = 3.14VTTWIQSFFHH516 pKa = 5.43QVVIFTQLAFVVLSYY531 pKa = 8.81FTTRR535 pKa = 11.84TVIQRR540 pKa = 11.84WDD542 pKa = 4.4LIPYY546 pKa = 9.31DD547 pKa = 3.81SRR549 pKa = 11.84RR550 pKa = 11.84SMVALTT556 pKa = 3.92
Molecular weight: 63.04 kDa
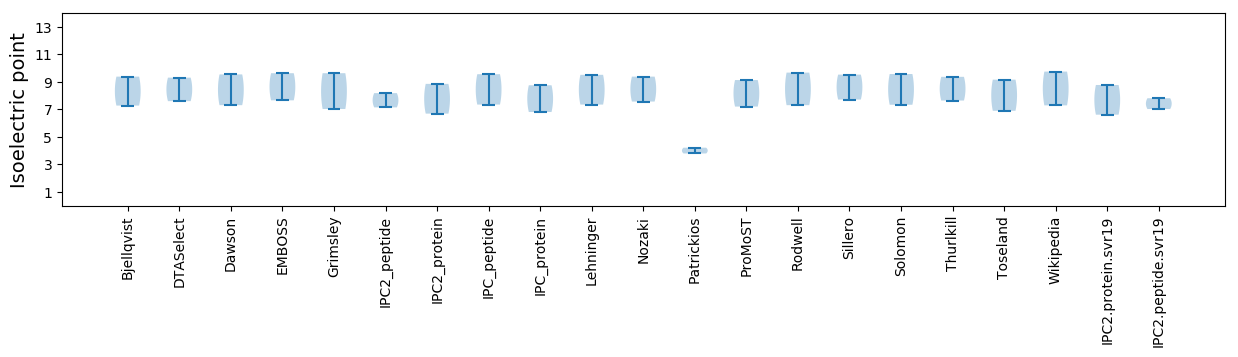
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGG9|A0A1L3KGG9_9VIRU RNA-directed RNA polymerase OS=Hubei diptera virus 15 OX=1922876 PE=4 SV=1
MM1 pKa = 7.57LRR3 pKa = 11.84RR4 pKa = 11.84FKK6 pKa = 11.03CRR8 pKa = 11.84VQPWSYY14 pKa = 11.58DD15 pKa = 3.47EE16 pKa = 5.22YY17 pKa = 10.58IEE19 pKa = 6.26SMDD22 pKa = 4.41CPSKK26 pKa = 10.59RR27 pKa = 11.84QFYY30 pKa = 10.91LDD32 pKa = 3.44TKK34 pKa = 11.21LDD36 pKa = 3.68MEE38 pKa = 4.91AGRR41 pKa = 11.84TISNRR46 pKa = 11.84ITPFTKK52 pKa = 10.27LEE54 pKa = 3.93KK55 pKa = 10.2MGTNKK60 pKa = 10.03YY61 pKa = 9.36KK62 pKa = 10.66APRR65 pKa = 11.84LIQARR70 pKa = 11.84HH71 pKa = 4.84PTFNLALGCYY81 pKa = 9.44IKK83 pKa = 10.61PLEE86 pKa = 4.32RR87 pKa = 11.84ALKK90 pKa = 10.4FRR92 pKa = 11.84DD93 pKa = 3.46NFGKK97 pKa = 8.96GTYY100 pKa = 9.25DD101 pKa = 3.25QIGAKK106 pKa = 8.12VARR109 pKa = 11.84LSRR112 pKa = 11.84KK113 pKa = 8.17YY114 pKa = 10.47KK115 pKa = 9.98YY116 pKa = 9.09YY117 pKa = 10.73TEE119 pKa = 4.92GDD121 pKa = 3.67HH122 pKa = 6.55LTFDD126 pKa = 3.34AHH128 pKa = 5.35VTVEE132 pKa = 3.93MLRR135 pKa = 11.84LCHH138 pKa = 6.74RR139 pKa = 11.84FYY141 pKa = 11.35LRR143 pKa = 11.84CFNNDD148 pKa = 2.7SKK150 pKa = 11.5LLRR153 pKa = 11.84LCGKK157 pKa = 7.5TIRR160 pKa = 11.84NRR162 pKa = 11.84ALSRR166 pKa = 11.84NGEE169 pKa = 4.14RR170 pKa = 11.84YY171 pKa = 6.95TVNGTRR177 pKa = 11.84MSGDD181 pKa = 3.0VDD183 pKa = 3.58TSFGNSIINYY193 pKa = 7.63YY194 pKa = 9.96IIRR197 pKa = 11.84SVLADD202 pKa = 3.41LHH204 pKa = 6.78LSGDD208 pKa = 4.2AIVNGDD214 pKa = 4.03DD215 pKa = 3.89YY216 pKa = 11.38IIFTDD221 pKa = 3.9VPIDD225 pKa = 3.74GSAATTAFRR234 pKa = 11.84RR235 pKa = 11.84YY236 pKa = 11.08NMDD239 pKa = 3.29TKK241 pKa = 11.1LLPSSSNIHH250 pKa = 5.1TVSFCRR256 pKa = 11.84TKK258 pKa = 10.88LFYY261 pKa = 10.92HH262 pKa = 7.31PDD264 pKa = 3.06GHH266 pKa = 6.37PTMGFDD272 pKa = 3.79PVRR275 pKa = 11.84LKK277 pKa = 10.76KK278 pKa = 10.41IYY280 pKa = 9.66GCTNIMYY287 pKa = 9.42PFEE290 pKa = 7.25DD291 pKa = 3.5YY292 pKa = 9.85MQYY295 pKa = 11.28LEE297 pKa = 5.51LIHH300 pKa = 6.52HH301 pKa = 7.37ANYY304 pKa = 9.28MININSPIHH313 pKa = 5.89TNWKK317 pKa = 8.93PLPKK321 pKa = 10.3LEE323 pKa = 4.55SKK325 pKa = 10.12LKK327 pKa = 10.53KK328 pKa = 10.58YY329 pKa = 7.05MTRR332 pKa = 11.84SLEE335 pKa = 4.1RR336 pKa = 11.84VFNKK340 pKa = 9.91QLSNKK345 pKa = 9.31PYY347 pKa = 10.29DD348 pKa = 4.01YY349 pKa = 11.0DD350 pKa = 3.95YY351 pKa = 10.08LTPSYY356 pKa = 10.75IEE358 pKa = 4.87AYY360 pKa = 9.01PDD362 pKa = 3.25YY363 pKa = 11.33VYY365 pKa = 11.06DD366 pKa = 3.66RR367 pKa = 11.84KK368 pKa = 11.25VNILYY373 pKa = 7.59TKK375 pKa = 9.01PKK377 pKa = 9.73PPIEE381 pKa = 4.42FIINHH386 pKa = 6.3NIKK389 pKa = 10.4EE390 pKa = 4.05ISQCC394 pKa = 3.74
MM1 pKa = 7.57LRR3 pKa = 11.84RR4 pKa = 11.84FKK6 pKa = 11.03CRR8 pKa = 11.84VQPWSYY14 pKa = 11.58DD15 pKa = 3.47EE16 pKa = 5.22YY17 pKa = 10.58IEE19 pKa = 6.26SMDD22 pKa = 4.41CPSKK26 pKa = 10.59RR27 pKa = 11.84QFYY30 pKa = 10.91LDD32 pKa = 3.44TKK34 pKa = 11.21LDD36 pKa = 3.68MEE38 pKa = 4.91AGRR41 pKa = 11.84TISNRR46 pKa = 11.84ITPFTKK52 pKa = 10.27LEE54 pKa = 3.93KK55 pKa = 10.2MGTNKK60 pKa = 10.03YY61 pKa = 9.36KK62 pKa = 10.66APRR65 pKa = 11.84LIQARR70 pKa = 11.84HH71 pKa = 4.84PTFNLALGCYY81 pKa = 9.44IKK83 pKa = 10.61PLEE86 pKa = 4.32RR87 pKa = 11.84ALKK90 pKa = 10.4FRR92 pKa = 11.84DD93 pKa = 3.46NFGKK97 pKa = 8.96GTYY100 pKa = 9.25DD101 pKa = 3.25QIGAKK106 pKa = 8.12VARR109 pKa = 11.84LSRR112 pKa = 11.84KK113 pKa = 8.17YY114 pKa = 10.47KK115 pKa = 9.98YY116 pKa = 9.09YY117 pKa = 10.73TEE119 pKa = 4.92GDD121 pKa = 3.67HH122 pKa = 6.55LTFDD126 pKa = 3.34AHH128 pKa = 5.35VTVEE132 pKa = 3.93MLRR135 pKa = 11.84LCHH138 pKa = 6.74RR139 pKa = 11.84FYY141 pKa = 11.35LRR143 pKa = 11.84CFNNDD148 pKa = 2.7SKK150 pKa = 11.5LLRR153 pKa = 11.84LCGKK157 pKa = 7.5TIRR160 pKa = 11.84NRR162 pKa = 11.84ALSRR166 pKa = 11.84NGEE169 pKa = 4.14RR170 pKa = 11.84YY171 pKa = 6.95TVNGTRR177 pKa = 11.84MSGDD181 pKa = 3.0VDD183 pKa = 3.58TSFGNSIINYY193 pKa = 7.63YY194 pKa = 9.96IIRR197 pKa = 11.84SVLADD202 pKa = 3.41LHH204 pKa = 6.78LSGDD208 pKa = 4.2AIVNGDD214 pKa = 4.03DD215 pKa = 3.89YY216 pKa = 11.38IIFTDD221 pKa = 3.9VPIDD225 pKa = 3.74GSAATTAFRR234 pKa = 11.84RR235 pKa = 11.84YY236 pKa = 11.08NMDD239 pKa = 3.29TKK241 pKa = 11.1LLPSSSNIHH250 pKa = 5.1TVSFCRR256 pKa = 11.84TKK258 pKa = 10.88LFYY261 pKa = 10.92HH262 pKa = 7.31PDD264 pKa = 3.06GHH266 pKa = 6.37PTMGFDD272 pKa = 3.79PVRR275 pKa = 11.84LKK277 pKa = 10.76KK278 pKa = 10.41IYY280 pKa = 9.66GCTNIMYY287 pKa = 9.42PFEE290 pKa = 7.25DD291 pKa = 3.5YY292 pKa = 9.85MQYY295 pKa = 11.28LEE297 pKa = 5.51LIHH300 pKa = 6.52HH301 pKa = 7.37ANYY304 pKa = 9.28MININSPIHH313 pKa = 5.89TNWKK317 pKa = 8.93PLPKK321 pKa = 10.3LEE323 pKa = 4.55SKK325 pKa = 10.12LKK327 pKa = 10.53KK328 pKa = 10.58YY329 pKa = 7.05MTRR332 pKa = 11.84SLEE335 pKa = 4.1RR336 pKa = 11.84VFNKK340 pKa = 9.91QLSNKK345 pKa = 9.31PYY347 pKa = 10.29DD348 pKa = 4.01YY349 pKa = 11.0DD350 pKa = 3.95YY351 pKa = 10.08LTPSYY356 pKa = 10.75IEE358 pKa = 4.87AYY360 pKa = 9.01PDD362 pKa = 3.25YY363 pKa = 11.33VYY365 pKa = 11.06DD366 pKa = 3.66RR367 pKa = 11.84KK368 pKa = 11.25VNILYY373 pKa = 7.59TKK375 pKa = 9.01PKK377 pKa = 9.73PPIEE381 pKa = 4.42FIINHH386 pKa = 6.3NIKK389 pKa = 10.4EE390 pKa = 4.05ISQCC394 pKa = 3.74
Molecular weight: 46.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
950 |
394 |
556 |
475.0 |
54.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.684 ± 0.987 | 1.684 ± 0.365 |
4.632 ± 1.042 | 4.316 ± 0.309 |
3.579 ± 0.602 | 3.158 ± 0.858 |
3.053 ± 0.004 | 7.158 ± 0.123 |
6.0 ± 0.981 | 10.0 ± 0.833 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.053 ± 0.004 | 4.421 ± 1.015 |
5.263 ± 0.041 | 3.684 ± 1.16 |
4.947 ± 1.467 | 8.211 ± 1.443 |
9.053 ± 1.492 | 4.737 ± 0.72 |
1.474 ± 0.587 | 5.895 ± 1.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
