
Mytilus coruscus (Sea mussel)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Mollusca; Bivalvia; Autobranchia; Pteriomorphia; Mytiloida; Mytiloidea; Mytilidae; Mytilinae; Mytilus
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
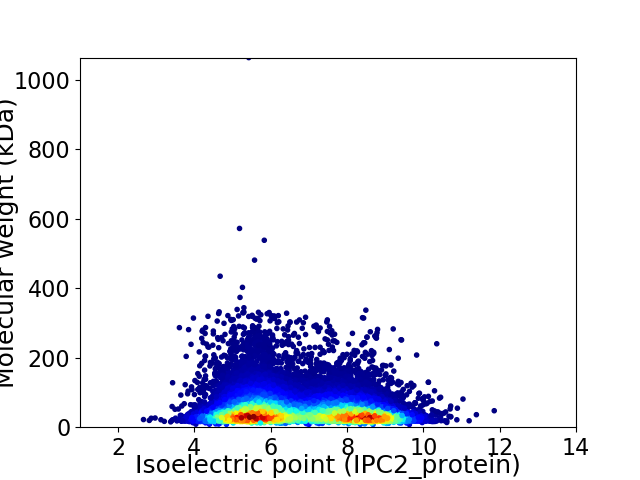
Virtual 2D-PAGE plot for 64248 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6J8B670|A0A6J8B670_MYTCO PLIN2 OS=Mytilus coruscus OX=42192 GN=MCOR_15481 PE=3 SV=1
MM1 pKa = 7.26VCDD4 pKa = 4.11LQVTTTLIRR13 pKa = 11.84DD14 pKa = 3.85FCDD17 pKa = 2.97TDD19 pKa = 3.23IDD21 pKa = 4.5GDD23 pKa = 4.2GVLNDD28 pKa = 3.92EE29 pKa = 5.0DD30 pKa = 3.91NCVFISNPSQTDD42 pKa = 2.51SDD44 pKa = 4.18GNHH47 pKa = 6.72RR48 pKa = 11.84GDD50 pKa = 3.54VCEE53 pKa = 4.77SDD55 pKa = 3.48YY56 pKa = 11.7DD57 pKa = 3.66QDD59 pKa = 3.92GTIDD63 pKa = 3.76TLDD66 pKa = 3.55NCPNNKK72 pKa = 9.84FINEE76 pKa = 4.23SDD78 pKa = 3.15FGTYY82 pKa = 9.14TGLDD86 pKa = 3.93LNPEE90 pKa = 4.26LTLQPAPSWMILHH103 pKa = 6.89GGKK106 pKa = 10.21EE107 pKa = 3.8IRR109 pKa = 11.84QVSVTTKK116 pKa = 10.02PVASVGNHH124 pKa = 6.16RR125 pKa = 11.84GDD127 pKa = 3.49VCEE130 pKa = 4.77SDD132 pKa = 3.48YY133 pKa = 11.7DD134 pKa = 3.66QDD136 pKa = 3.92GTIDD140 pKa = 3.76TLDD143 pKa = 3.55NCPNNKK149 pKa = 9.84FINEE153 pKa = 3.93SDD155 pKa = 3.25FGYY158 pKa = 10.53LYY160 pKa = 10.35WFRR163 pKa = 11.84PKK165 pKa = 10.6SS166 pKa = 3.43
MM1 pKa = 7.26VCDD4 pKa = 4.11LQVTTTLIRR13 pKa = 11.84DD14 pKa = 3.85FCDD17 pKa = 2.97TDD19 pKa = 3.23IDD21 pKa = 4.5GDD23 pKa = 4.2GVLNDD28 pKa = 3.92EE29 pKa = 5.0DD30 pKa = 3.91NCVFISNPSQTDD42 pKa = 2.51SDD44 pKa = 4.18GNHH47 pKa = 6.72RR48 pKa = 11.84GDD50 pKa = 3.54VCEE53 pKa = 4.77SDD55 pKa = 3.48YY56 pKa = 11.7DD57 pKa = 3.66QDD59 pKa = 3.92GTIDD63 pKa = 3.76TLDD66 pKa = 3.55NCPNNKK72 pKa = 9.84FINEE76 pKa = 4.23SDD78 pKa = 3.15FGTYY82 pKa = 9.14TGLDD86 pKa = 3.93LNPEE90 pKa = 4.26LTLQPAPSWMILHH103 pKa = 6.89GGKK106 pKa = 10.21EE107 pKa = 3.8IRR109 pKa = 11.84QVSVTTKK116 pKa = 10.02PVASVGNHH124 pKa = 6.16RR125 pKa = 11.84GDD127 pKa = 3.49VCEE130 pKa = 4.77SDD132 pKa = 3.48YY133 pKa = 11.7DD134 pKa = 3.66QDD136 pKa = 3.92GTIDD140 pKa = 3.76TLDD143 pKa = 3.55NCPNNKK149 pKa = 9.84FINEE153 pKa = 3.93SDD155 pKa = 3.25FGYY158 pKa = 10.53LYY160 pKa = 10.35WFRR163 pKa = 11.84PKK165 pKa = 10.6SS166 pKa = 3.43
Molecular weight: 18.49 kDa
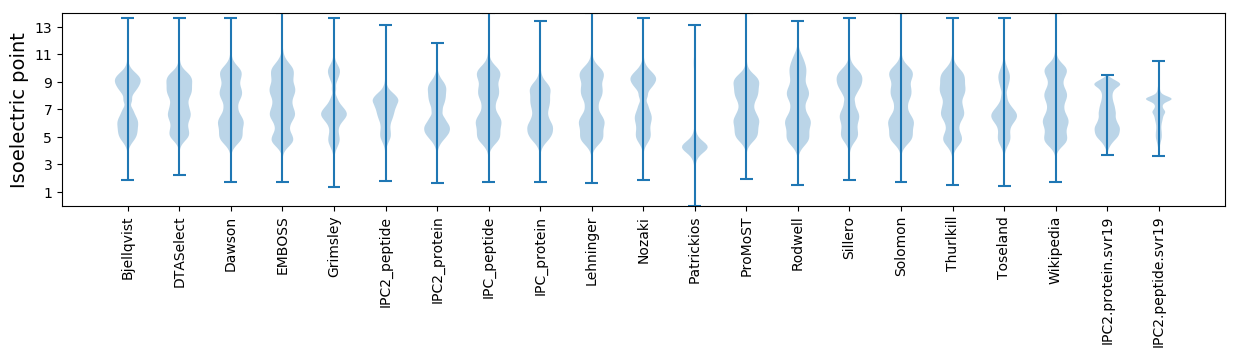
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6J8EZ31|A0A6J8EZ31_MYTCO G_PROTEIN_RECEP_F1_2 domain-containing protein OS=Mytilus coruscus OX=42192 GN=MCOR_57689 PE=3 SV=1
MM1 pKa = 6.96YY2 pKa = 10.27LAYY5 pKa = 10.19NSNGIRR11 pKa = 11.84TPVGGRR17 pKa = 11.84VINKK21 pKa = 5.57QTKK24 pKa = 9.2HH25 pKa = 5.37SKK27 pKa = 10.23QSITHH32 pKa = 6.88LSSQQSITHH41 pKa = 6.89LSRR44 pKa = 11.84VINNQTKK51 pKa = 9.85HH52 pKa = 5.22RR53 pKa = 11.84QQSITHH59 pKa = 6.93LSRR62 pKa = 11.84VINKK66 pKa = 6.69QTKK69 pKa = 9.13HH70 pKa = 5.14RR71 pKa = 11.84QQSITHH77 pKa = 6.93LSRR80 pKa = 11.84VINKK84 pKa = 6.69QTKK87 pKa = 9.13HH88 pKa = 5.14RR89 pKa = 11.84QQSITHH95 pKa = 6.93LSRR98 pKa = 11.84VINKK102 pKa = 6.69QTKK105 pKa = 9.13HH106 pKa = 5.14RR107 pKa = 11.84QQSITHH113 pKa = 6.93LSRR116 pKa = 11.84VINKK120 pKa = 6.88QTKK123 pKa = 8.56HH124 pKa = 5.29HH125 pKa = 5.48QQSITHH131 pKa = 7.05LSRR134 pKa = 11.84VINKK138 pKa = 6.69QTKK141 pKa = 9.13HH142 pKa = 5.14RR143 pKa = 11.84QQSITHH149 pKa = 6.92LSRR152 pKa = 11.84VINNQTKK159 pKa = 10.15HH160 pKa = 4.98SQQSITHH167 pKa = 6.88LSRR170 pKa = 11.84VINKK174 pKa = 6.86QTKK177 pKa = 9.52HH178 pKa = 4.93SQQSITHH185 pKa = 6.88LSRR188 pKa = 11.84VINKK192 pKa = 6.69QTKK195 pKa = 9.13HH196 pKa = 5.14RR197 pKa = 11.84QQSITHH203 pKa = 6.93LSRR206 pKa = 11.84VINKK210 pKa = 6.67QTKK213 pKa = 9.2HH214 pKa = 5.37SKK216 pKa = 10.23QSITHH221 pKa = 6.91LSRR224 pKa = 11.84IINKK228 pKa = 6.6QTKK231 pKa = 9.12HH232 pKa = 5.15RR233 pKa = 11.84QQSITHH239 pKa = 6.93LSRR242 pKa = 11.84VINKK246 pKa = 6.67QTKK249 pKa = 9.2HH250 pKa = 5.37SKK252 pKa = 10.23QSITHH257 pKa = 6.79LSRR260 pKa = 11.84VINKK264 pKa = 6.69QTKK267 pKa = 9.13HH268 pKa = 5.14RR269 pKa = 11.84QQSITHH275 pKa = 6.93LSRR278 pKa = 11.84VINKK282 pKa = 6.69QTKK285 pKa = 9.13HH286 pKa = 5.14RR287 pKa = 11.84QQSITHH293 pKa = 6.93LSRR296 pKa = 11.84VINKK300 pKa = 6.77QTKK303 pKa = 8.19HH304 pKa = 6.31HH305 pKa = 6.24II306 pKa = 3.92
MM1 pKa = 6.96YY2 pKa = 10.27LAYY5 pKa = 10.19NSNGIRR11 pKa = 11.84TPVGGRR17 pKa = 11.84VINKK21 pKa = 5.57QTKK24 pKa = 9.2HH25 pKa = 5.37SKK27 pKa = 10.23QSITHH32 pKa = 6.88LSSQQSITHH41 pKa = 6.89LSRR44 pKa = 11.84VINNQTKK51 pKa = 9.85HH52 pKa = 5.22RR53 pKa = 11.84QQSITHH59 pKa = 6.93LSRR62 pKa = 11.84VINKK66 pKa = 6.69QTKK69 pKa = 9.13HH70 pKa = 5.14RR71 pKa = 11.84QQSITHH77 pKa = 6.93LSRR80 pKa = 11.84VINKK84 pKa = 6.69QTKK87 pKa = 9.13HH88 pKa = 5.14RR89 pKa = 11.84QQSITHH95 pKa = 6.93LSRR98 pKa = 11.84VINKK102 pKa = 6.69QTKK105 pKa = 9.13HH106 pKa = 5.14RR107 pKa = 11.84QQSITHH113 pKa = 6.93LSRR116 pKa = 11.84VINKK120 pKa = 6.88QTKK123 pKa = 8.56HH124 pKa = 5.29HH125 pKa = 5.48QQSITHH131 pKa = 7.05LSRR134 pKa = 11.84VINKK138 pKa = 6.69QTKK141 pKa = 9.13HH142 pKa = 5.14RR143 pKa = 11.84QQSITHH149 pKa = 6.92LSRR152 pKa = 11.84VINNQTKK159 pKa = 10.15HH160 pKa = 4.98SQQSITHH167 pKa = 6.88LSRR170 pKa = 11.84VINKK174 pKa = 6.86QTKK177 pKa = 9.52HH178 pKa = 4.93SQQSITHH185 pKa = 6.88LSRR188 pKa = 11.84VINKK192 pKa = 6.69QTKK195 pKa = 9.13HH196 pKa = 5.14RR197 pKa = 11.84QQSITHH203 pKa = 6.93LSRR206 pKa = 11.84VINKK210 pKa = 6.67QTKK213 pKa = 9.2HH214 pKa = 5.37SKK216 pKa = 10.23QSITHH221 pKa = 6.91LSRR224 pKa = 11.84IINKK228 pKa = 6.6QTKK231 pKa = 9.12HH232 pKa = 5.15RR233 pKa = 11.84QQSITHH239 pKa = 6.93LSRR242 pKa = 11.84VINKK246 pKa = 6.67QTKK249 pKa = 9.2HH250 pKa = 5.37SKK252 pKa = 10.23QSITHH257 pKa = 6.79LSRR260 pKa = 11.84VINKK264 pKa = 6.69QTKK267 pKa = 9.13HH268 pKa = 5.14RR269 pKa = 11.84QQSITHH275 pKa = 6.93LSRR278 pKa = 11.84VINKK282 pKa = 6.69QTKK285 pKa = 9.13HH286 pKa = 5.14RR287 pKa = 11.84QQSITHH293 pKa = 6.93LSRR296 pKa = 11.84VINKK300 pKa = 6.77QTKK303 pKa = 8.19HH304 pKa = 6.31HH305 pKa = 6.24II306 pKa = 3.92
Molecular weight: 36.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
29520201 |
66 |
9566 |
459.5 |
52.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.69 ± 0.008 | 2.365 ± 0.009 |
5.994 ± 0.007 | 6.508 ± 0.012 |
3.921 ± 0.006 | 5.099 ± 0.012 |
2.524 ± 0.005 | 6.532 ± 0.009 |
7.543 ± 0.013 | 8.393 ± 0.011 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.341 ± 0.005 | 5.861 ± 0.009 |
4.184 ± 0.009 | 4.333 ± 0.008 |
4.757 ± 0.008 | 8.157 ± 0.012 |
6.38 ± 0.014 | 5.976 ± 0.008 |
1.044 ± 0.003 | 3.398 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
