
Clostridium vincentii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
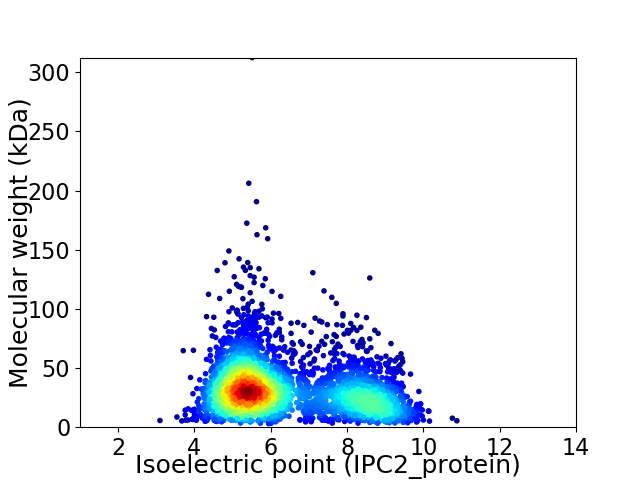
Virtual 2D-PAGE plot for 3379 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T0BI67|A0A2T0BI67_9CLOT Glucans biosynthesis protein C OS=Clostridium vincentii OX=52704 GN=mdoC PE=4 SV=1
MM1 pKa = 7.04YY2 pKa = 9.94FKK4 pKa = 11.11LINEE8 pKa = 4.04KK9 pKa = 9.67HH10 pKa = 5.86AKK12 pKa = 9.07IVACIAIVCALLFTVTILFIPKK34 pKa = 9.65NVSDD38 pKa = 4.55DD39 pKa = 3.87ASGTAAYY46 pKa = 10.75SDD48 pKa = 3.54TLFNEE53 pKa = 4.97DD54 pKa = 4.13EE55 pKa = 4.33VSDD58 pKa = 3.83IQISIDD64 pKa = 3.61DD65 pKa = 4.4EE66 pKa = 4.44SWASILEE73 pKa = 4.23SPLDD77 pKa = 3.57EE78 pKa = 4.82TYY80 pKa = 10.71YY81 pKa = 11.12SCDD84 pKa = 2.62ITINGEE90 pKa = 3.71TLYY93 pKa = 11.18NVGIRR98 pKa = 11.84PKK100 pKa = 10.79GNTSLTQVASSDD112 pKa = 3.58SEE114 pKa = 4.18RR115 pKa = 11.84YY116 pKa = 8.73SFKK119 pKa = 11.24VKK121 pKa = 10.18FDD123 pKa = 3.91EE124 pKa = 4.51YY125 pKa = 11.58VDD127 pKa = 3.64GQTYY131 pKa = 10.97NGLDD135 pKa = 3.52KK136 pKa = 11.55LNLNNTYY143 pKa = 11.1SDD145 pKa = 3.07ATYY148 pKa = 9.78LKK150 pKa = 10.22EE151 pKa = 4.16YY152 pKa = 10.29ISYY155 pKa = 11.22DD156 pKa = 3.25LFDD159 pKa = 4.54FMGVTTPEE167 pKa = 4.05TAFSNITINSEE178 pKa = 3.63NRR180 pKa = 11.84GVYY183 pKa = 10.02LAVEE187 pKa = 4.28GLEE190 pKa = 3.87EE191 pKa = 4.73SYY193 pKa = 11.21LSRR196 pKa = 11.84NYY198 pKa = 10.87GSDD201 pKa = 2.93SGNLYY206 pKa = 9.98KK207 pKa = 10.71AEE209 pKa = 4.11GTGTDD214 pKa = 4.23LVWNGDD220 pKa = 3.63TQSNYY225 pKa = 10.9SGLKK229 pKa = 9.98DD230 pKa = 3.73NSVKK234 pKa = 10.84DD235 pKa = 3.35ITDD238 pKa = 3.2EE239 pKa = 4.17AFQKK243 pKa = 10.47VIDD246 pKa = 4.6MISNLNEE253 pKa = 3.67GTNLEE258 pKa = 4.51DD259 pKa = 4.5YY260 pKa = 10.89IDD262 pKa = 3.95VEE264 pKa = 4.19ATLKK268 pKa = 10.67YY269 pKa = 10.05FAVSTALVNLDD280 pKa = 4.05SYY282 pKa = 11.51QCNLYY287 pKa = 9.85HH288 pKa = 6.96NYY290 pKa = 10.39YY291 pKa = 9.86IYY293 pKa = 10.88EE294 pKa = 4.11EE295 pKa = 5.31DD296 pKa = 4.18GVCTILPWDD305 pKa = 4.06LNLSFGAFSGGGGGNNARR323 pKa = 11.84NNNDD327 pKa = 3.08GDD329 pKa = 3.99NATTTTNTTRR339 pKa = 11.84GIINFPIDD347 pKa = 3.87EE348 pKa = 4.52PTSATLEE355 pKa = 4.18DD356 pKa = 4.35SPLLSKK362 pKa = 10.82LLEE365 pKa = 4.0VDD367 pKa = 3.87EE368 pKa = 4.64YY369 pKa = 11.58KK370 pKa = 11.2DD371 pKa = 3.46MYY373 pKa = 10.77HH374 pKa = 7.66DD375 pKa = 3.92YY376 pKa = 11.26LNTIVEE382 pKa = 4.72EE383 pKa = 4.44YY384 pKa = 10.17FDD386 pKa = 4.2SGTFEE391 pKa = 4.76SKK393 pKa = 11.19VNTTSTLIDD402 pKa = 3.51SYY404 pKa = 11.88VEE406 pKa = 3.96NNPTAFYY413 pKa = 10.25GYY415 pKa = 9.88DD416 pKa = 3.28QFSASIDD423 pKa = 3.6EE424 pKa = 4.65LSTFGTYY431 pKa = 8.82RR432 pKa = 11.84AKK434 pKa = 10.59SIAMQLSGEE443 pKa = 4.17IPSTKK448 pKa = 10.1AGQSEE453 pKa = 4.55VDD455 pKa = 3.2LTTYY459 pKa = 10.61FSEE462 pKa = 5.61DD463 pKa = 3.61SVDD466 pKa = 3.88MNILGSMGGGNKK478 pKa = 10.47GGDD481 pKa = 3.26QGNNTDD487 pKa = 3.56TGTVAEE493 pKa = 4.54GQMPAGFPSGDD504 pKa = 3.55MDD506 pKa = 4.0QGGANMTPSSDD517 pKa = 3.46VQQNGSPNTDD527 pKa = 3.01TTKK530 pKa = 10.58SEE532 pKa = 4.09RR533 pKa = 11.84DD534 pKa = 3.72DD535 pKa = 5.12NGMQAPDD542 pKa = 4.26GNNMTRR548 pKa = 11.84PDD550 pKa = 3.85GMSKK554 pKa = 11.12GDD556 pKa = 3.69TNTSISITSIIIFATSFISIIIALVFSIKK585 pKa = 10.35FKK587 pKa = 10.62RR588 pKa = 11.84KK589 pKa = 9.39KK590 pKa = 10.44FKK592 pKa = 10.85LL593 pKa = 3.56
MM1 pKa = 7.04YY2 pKa = 9.94FKK4 pKa = 11.11LINEE8 pKa = 4.04KK9 pKa = 9.67HH10 pKa = 5.86AKK12 pKa = 9.07IVACIAIVCALLFTVTILFIPKK34 pKa = 9.65NVSDD38 pKa = 4.55DD39 pKa = 3.87ASGTAAYY46 pKa = 10.75SDD48 pKa = 3.54TLFNEE53 pKa = 4.97DD54 pKa = 4.13EE55 pKa = 4.33VSDD58 pKa = 3.83IQISIDD64 pKa = 3.61DD65 pKa = 4.4EE66 pKa = 4.44SWASILEE73 pKa = 4.23SPLDD77 pKa = 3.57EE78 pKa = 4.82TYY80 pKa = 10.71YY81 pKa = 11.12SCDD84 pKa = 2.62ITINGEE90 pKa = 3.71TLYY93 pKa = 11.18NVGIRR98 pKa = 11.84PKK100 pKa = 10.79GNTSLTQVASSDD112 pKa = 3.58SEE114 pKa = 4.18RR115 pKa = 11.84YY116 pKa = 8.73SFKK119 pKa = 11.24VKK121 pKa = 10.18FDD123 pKa = 3.91EE124 pKa = 4.51YY125 pKa = 11.58VDD127 pKa = 3.64GQTYY131 pKa = 10.97NGLDD135 pKa = 3.52KK136 pKa = 11.55LNLNNTYY143 pKa = 11.1SDD145 pKa = 3.07ATYY148 pKa = 9.78LKK150 pKa = 10.22EE151 pKa = 4.16YY152 pKa = 10.29ISYY155 pKa = 11.22DD156 pKa = 3.25LFDD159 pKa = 4.54FMGVTTPEE167 pKa = 4.05TAFSNITINSEE178 pKa = 3.63NRR180 pKa = 11.84GVYY183 pKa = 10.02LAVEE187 pKa = 4.28GLEE190 pKa = 3.87EE191 pKa = 4.73SYY193 pKa = 11.21LSRR196 pKa = 11.84NYY198 pKa = 10.87GSDD201 pKa = 2.93SGNLYY206 pKa = 9.98KK207 pKa = 10.71AEE209 pKa = 4.11GTGTDD214 pKa = 4.23LVWNGDD220 pKa = 3.63TQSNYY225 pKa = 10.9SGLKK229 pKa = 9.98DD230 pKa = 3.73NSVKK234 pKa = 10.84DD235 pKa = 3.35ITDD238 pKa = 3.2EE239 pKa = 4.17AFQKK243 pKa = 10.47VIDD246 pKa = 4.6MISNLNEE253 pKa = 3.67GTNLEE258 pKa = 4.51DD259 pKa = 4.5YY260 pKa = 10.89IDD262 pKa = 3.95VEE264 pKa = 4.19ATLKK268 pKa = 10.67YY269 pKa = 10.05FAVSTALVNLDD280 pKa = 4.05SYY282 pKa = 11.51QCNLYY287 pKa = 9.85HH288 pKa = 6.96NYY290 pKa = 10.39YY291 pKa = 9.86IYY293 pKa = 10.88EE294 pKa = 4.11EE295 pKa = 5.31DD296 pKa = 4.18GVCTILPWDD305 pKa = 4.06LNLSFGAFSGGGGGNNARR323 pKa = 11.84NNNDD327 pKa = 3.08GDD329 pKa = 3.99NATTTTNTTRR339 pKa = 11.84GIINFPIDD347 pKa = 3.87EE348 pKa = 4.52PTSATLEE355 pKa = 4.18DD356 pKa = 4.35SPLLSKK362 pKa = 10.82LLEE365 pKa = 4.0VDD367 pKa = 3.87EE368 pKa = 4.64YY369 pKa = 11.58KK370 pKa = 11.2DD371 pKa = 3.46MYY373 pKa = 10.77HH374 pKa = 7.66DD375 pKa = 3.92YY376 pKa = 11.26LNTIVEE382 pKa = 4.72EE383 pKa = 4.44YY384 pKa = 10.17FDD386 pKa = 4.2SGTFEE391 pKa = 4.76SKK393 pKa = 11.19VNTTSTLIDD402 pKa = 3.51SYY404 pKa = 11.88VEE406 pKa = 3.96NNPTAFYY413 pKa = 10.25GYY415 pKa = 9.88DD416 pKa = 3.28QFSASIDD423 pKa = 3.6EE424 pKa = 4.65LSTFGTYY431 pKa = 8.82RR432 pKa = 11.84AKK434 pKa = 10.59SIAMQLSGEE443 pKa = 4.17IPSTKK448 pKa = 10.1AGQSEE453 pKa = 4.55VDD455 pKa = 3.2LTTYY459 pKa = 10.61FSEE462 pKa = 5.61DD463 pKa = 3.61SVDD466 pKa = 3.88MNILGSMGGGNKK478 pKa = 10.47GGDD481 pKa = 3.26QGNNTDD487 pKa = 3.56TGTVAEE493 pKa = 4.54GQMPAGFPSGDD504 pKa = 3.55MDD506 pKa = 4.0QGGANMTPSSDD517 pKa = 3.46VQQNGSPNTDD527 pKa = 3.01TTKK530 pKa = 10.58SEE532 pKa = 4.09RR533 pKa = 11.84DD534 pKa = 3.72DD535 pKa = 5.12NGMQAPDD542 pKa = 4.26GNNMTRR548 pKa = 11.84PDD550 pKa = 3.85GMSKK554 pKa = 11.12GDD556 pKa = 3.69TNTSISITSIIIFATSFISIIIALVFSIKK585 pKa = 10.35FKK587 pKa = 10.62RR588 pKa = 11.84KK589 pKa = 9.39KK590 pKa = 10.44FKK592 pKa = 10.85LL593 pKa = 3.56
Molecular weight: 65.02 kDa
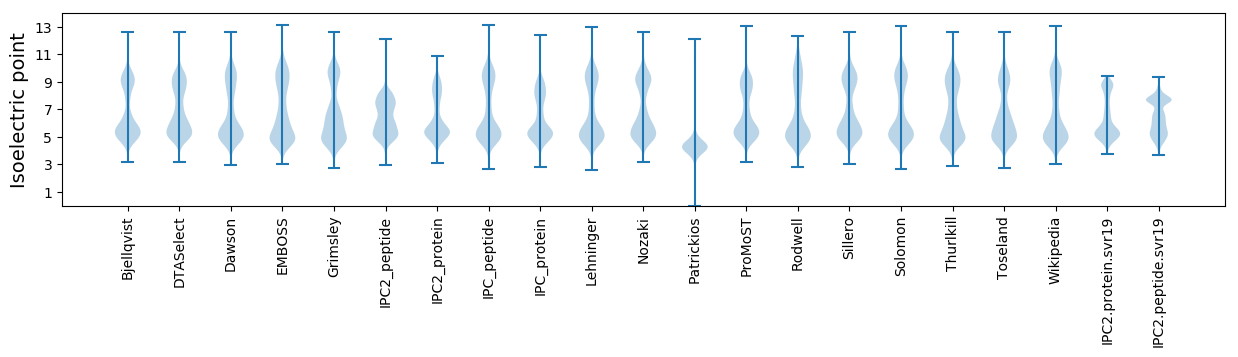
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T0BBW7|A0A2T0BBW7_9CLOT Putative membrane protein insertion efficiency factor OS=Clostridium vincentii OX=52704 GN=CLVI_25140 PE=3 SV=1
MM1 pKa = 7.56FMTFQPKK8 pKa = 8.99KK9 pKa = 7.62RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.25RR14 pKa = 11.84EE15 pKa = 3.66HH16 pKa = 6.22GFRR19 pKa = 11.84KK20 pKa = 10.06RR21 pKa = 11.84MATPAGRR28 pKa = 11.84NILRR32 pKa = 11.84MRR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.57GRR39 pKa = 11.84KK40 pKa = 8.93KK41 pKa = 9.66LTAA44 pKa = 4.2
MM1 pKa = 7.56FMTFQPKK8 pKa = 8.99KK9 pKa = 7.62RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.25RR14 pKa = 11.84EE15 pKa = 3.66HH16 pKa = 6.22GFRR19 pKa = 11.84KK20 pKa = 10.06RR21 pKa = 11.84MATPAGRR28 pKa = 11.84NILRR32 pKa = 11.84MRR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.57GRR39 pKa = 11.84KK40 pKa = 8.93KK41 pKa = 9.66LTAA44 pKa = 4.2
Molecular weight: 5.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1015978 |
29 |
2723 |
300.7 |
33.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.654 ± 0.036 | 1.185 ± 0.014 |
5.606 ± 0.03 | 7.532 ± 0.045 |
4.366 ± 0.033 | 6.576 ± 0.038 |
1.323 ± 0.016 | 10.175 ± 0.05 |
8.831 ± 0.033 | 9.118 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.662 ± 0.02 | 6.036 ± 0.04 |
2.705 ± 0.024 | 2.383 ± 0.023 |
3.327 ± 0.026 | 6.39 ± 0.034 |
5.034 ± 0.031 | 6.437 ± 0.029 |
0.674 ± 0.014 | 3.987 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
