
Aspergillus ustus
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
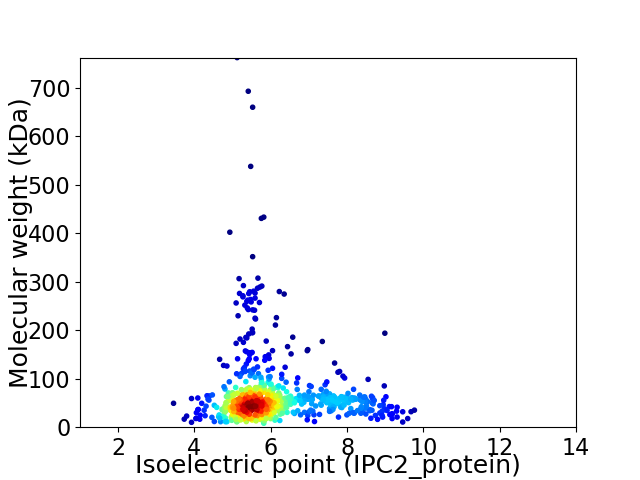
Virtual 2D-PAGE plot for 706 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
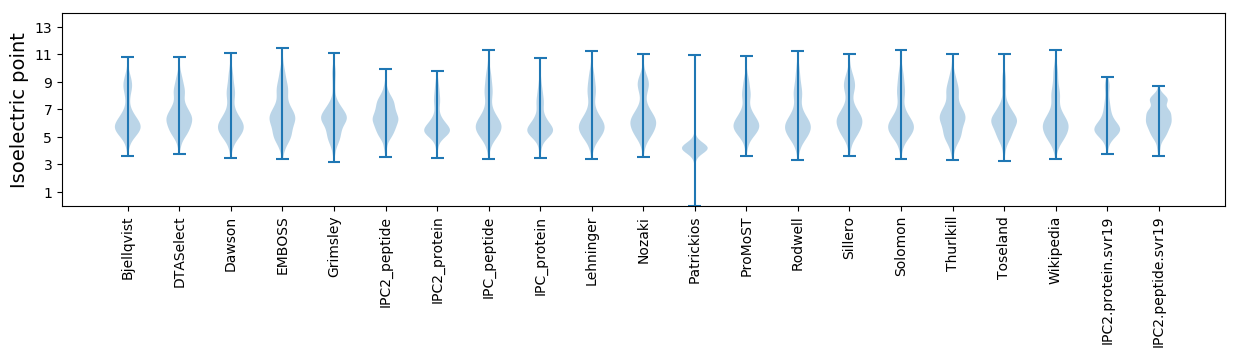
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C1E646|A0A0C1E646_ASPUT C6 zinc finger domain-containing protein OS=Aspergillus ustus OX=40382 GN=HK57_00612 PE=4 SV=1
MM1 pKa = 7.66RR2 pKa = 11.84FSVLAFIALTASVMASDD19 pKa = 5.54DD20 pKa = 4.13PGPSPTEE27 pKa = 4.06SIGCEE32 pKa = 3.46PHH34 pKa = 6.72GDD36 pKa = 3.33HH37 pKa = 6.46WHH39 pKa = 7.11CDD41 pKa = 3.34APATSTLATTTSSTSEE57 pKa = 3.89SDD59 pKa = 3.43EE60 pKa = 4.85DD61 pKa = 4.46DD62 pKa = 4.06SSVTPGPSPTEE73 pKa = 4.12SIGCEE78 pKa = 3.46PHH80 pKa = 6.57GDD82 pKa = 3.25HH83 pKa = 6.39WHH85 pKa = 6.9CEE87 pKa = 3.79AAATDD92 pKa = 3.49VDD94 pKa = 4.34EE95 pKa = 5.13EE96 pKa = 4.82DD97 pKa = 4.01EE98 pKa = 4.6SVTPGPSPTEE108 pKa = 4.12SIGCEE113 pKa = 3.46PHH115 pKa = 6.72GDD117 pKa = 3.33HH118 pKa = 6.46WHH120 pKa = 7.11CDD122 pKa = 3.36APATTTTDD130 pKa = 3.12SDD132 pKa = 4.43SEE134 pKa = 4.64SSTSAAADD142 pKa = 3.69EE143 pKa = 5.27DD144 pKa = 4.56SSSVTPPPSPTEE156 pKa = 4.04SVGCEE161 pKa = 3.43PHH163 pKa = 6.8GDD165 pKa = 3.13HH166 pKa = 6.47WHH168 pKa = 6.95CDD170 pKa = 3.55GPRR173 pKa = 11.84EE174 pKa = 4.0TDD176 pKa = 3.03EE177 pKa = 4.27ATEE180 pKa = 4.22SSSNATSTTEE190 pKa = 3.46EE191 pKa = 4.23GAAEE195 pKa = 4.0EE196 pKa = 4.41TGDD199 pKa = 4.76DD200 pKa = 3.79EE201 pKa = 4.82DD202 pKa = 3.91AAGIVGVKK210 pKa = 10.27LSGVVAVAVAAAAALLL226 pKa = 4.0
MM1 pKa = 7.66RR2 pKa = 11.84FSVLAFIALTASVMASDD19 pKa = 5.54DD20 pKa = 4.13PGPSPTEE27 pKa = 4.06SIGCEE32 pKa = 3.46PHH34 pKa = 6.72GDD36 pKa = 3.33HH37 pKa = 6.46WHH39 pKa = 7.11CDD41 pKa = 3.34APATSTLATTTSSTSEE57 pKa = 3.89SDD59 pKa = 3.43EE60 pKa = 4.85DD61 pKa = 4.46DD62 pKa = 4.06SSVTPGPSPTEE73 pKa = 4.12SIGCEE78 pKa = 3.46PHH80 pKa = 6.57GDD82 pKa = 3.25HH83 pKa = 6.39WHH85 pKa = 6.9CEE87 pKa = 3.79AAATDD92 pKa = 3.49VDD94 pKa = 4.34EE95 pKa = 5.13EE96 pKa = 4.82DD97 pKa = 4.01EE98 pKa = 4.6SVTPGPSPTEE108 pKa = 4.12SIGCEE113 pKa = 3.46PHH115 pKa = 6.72GDD117 pKa = 3.33HH118 pKa = 6.46WHH120 pKa = 7.11CDD122 pKa = 3.36APATTTTDD130 pKa = 3.12SDD132 pKa = 4.43SEE134 pKa = 4.64SSTSAAADD142 pKa = 3.69EE143 pKa = 5.27DD144 pKa = 4.56SSSVTPPPSPTEE156 pKa = 4.04SVGCEE161 pKa = 3.43PHH163 pKa = 6.8GDD165 pKa = 3.13HH166 pKa = 6.47WHH168 pKa = 6.95CDD170 pKa = 3.55GPRR173 pKa = 11.84EE174 pKa = 4.0TDD176 pKa = 3.03EE177 pKa = 4.27ATEE180 pKa = 4.22SSSNATSTTEE190 pKa = 3.46EE191 pKa = 4.23GAAEE195 pKa = 4.0EE196 pKa = 4.41TGDD199 pKa = 4.76DD200 pKa = 3.79EE201 pKa = 4.82DD202 pKa = 3.91AAGIVGVKK210 pKa = 10.27LSGVVAVAVAAAAALLL226 pKa = 4.0
Molecular weight: 22.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C1E790|A0A0C1E790_ASPUT Integral membrane protein OS=Aspergillus ustus OX=40382 GN=HK57_00165 PE=4 SV=1
MM1 pKa = 7.59SRR3 pKa = 11.84GLSALHH9 pKa = 6.39SLLPTVEE16 pKa = 4.03KK17 pKa = 10.75KK18 pKa = 9.88KK19 pKa = 10.68AKK21 pKa = 9.93KK22 pKa = 9.45PRR24 pKa = 11.84PLHH27 pKa = 6.07TIGTSKK33 pKa = 10.69KK34 pKa = 9.32NPIRR38 pKa = 11.84QDD40 pKa = 3.42EE41 pKa = 4.3PPIQCRR47 pKa = 11.84YY48 pKa = 6.47NTHH51 pKa = 5.66AHH53 pKa = 3.93VHH55 pKa = 5.63IPINPSPSSRR65 pKa = 11.84ASTRR69 pKa = 11.84FLLSLRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84PRR80 pKa = 11.84RR81 pKa = 11.84PARR84 pKa = 11.84LPRR87 pKa = 11.84SRR89 pKa = 11.84AHH91 pKa = 7.07LPGTLPRR98 pKa = 11.84PIKK101 pKa = 10.23ARR103 pKa = 11.84HH104 pKa = 5.91LRR106 pKa = 11.84AQYY109 pKa = 9.63PSTLIVHH116 pKa = 7.3RR117 pKa = 11.84GNAHH121 pKa = 6.0NVTDD125 pKa = 3.98VLACLTVPNSSVRR138 pKa = 11.84QLVSAITFTIGNKK151 pKa = 9.38PDD153 pKa = 3.63LKK155 pKa = 11.19KK156 pKa = 10.38MGEE159 pKa = 4.21GDD161 pKa = 3.83PLVCQKK167 pKa = 10.62GMAVLLEE174 pKa = 4.28ALSVLRR180 pKa = 11.84RR181 pKa = 11.84DD182 pKa = 3.72WGVHH186 pKa = 4.73GQPLLCVVSTTGISEE201 pKa = 4.17TRR203 pKa = 11.84DD204 pKa = 3.16IPLLFYY210 pKa = 10.44PLYY213 pKa = 9.04HH214 pKa = 6.62TLLAVPHH221 pKa = 6.75RR222 pKa = 11.84DD223 pKa = 3.27KK224 pKa = 11.39KK225 pKa = 10.85VMEE228 pKa = 4.48GLLVASGEE236 pKa = 4.17RR237 pKa = 11.84FVLVRR242 pKa = 11.84PSLLWDD248 pKa = 3.51GEE250 pKa = 4.15RR251 pKa = 11.84RR252 pKa = 11.84KK253 pKa = 10.86GKK255 pKa = 10.51GKK257 pKa = 9.96GNGVVRR263 pKa = 11.84VGIADD268 pKa = 3.27ARR270 pKa = 11.84TGVVEE275 pKa = 4.28RR276 pKa = 11.84KK277 pKa = 9.18EE278 pKa = 3.82VGYY281 pKa = 8.78TISRR285 pKa = 11.84EE286 pKa = 4.13AVGGWMFEE294 pKa = 3.89HH295 pKa = 7.31LILRR299 pKa = 11.84AGEE302 pKa = 3.95KK303 pKa = 10.76GLVFEE308 pKa = 5.05EE309 pKa = 4.73KK310 pKa = 10.4AVSLTWW316 pKa = 3.56
MM1 pKa = 7.59SRR3 pKa = 11.84GLSALHH9 pKa = 6.39SLLPTVEE16 pKa = 4.03KK17 pKa = 10.75KK18 pKa = 9.88KK19 pKa = 10.68AKK21 pKa = 9.93KK22 pKa = 9.45PRR24 pKa = 11.84PLHH27 pKa = 6.07TIGTSKK33 pKa = 10.69KK34 pKa = 9.32NPIRR38 pKa = 11.84QDD40 pKa = 3.42EE41 pKa = 4.3PPIQCRR47 pKa = 11.84YY48 pKa = 6.47NTHH51 pKa = 5.66AHH53 pKa = 3.93VHH55 pKa = 5.63IPINPSPSSRR65 pKa = 11.84ASTRR69 pKa = 11.84FLLSLRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84PRR80 pKa = 11.84RR81 pKa = 11.84PARR84 pKa = 11.84LPRR87 pKa = 11.84SRR89 pKa = 11.84AHH91 pKa = 7.07LPGTLPRR98 pKa = 11.84PIKK101 pKa = 10.23ARR103 pKa = 11.84HH104 pKa = 5.91LRR106 pKa = 11.84AQYY109 pKa = 9.63PSTLIVHH116 pKa = 7.3RR117 pKa = 11.84GNAHH121 pKa = 6.0NVTDD125 pKa = 3.98VLACLTVPNSSVRR138 pKa = 11.84QLVSAITFTIGNKK151 pKa = 9.38PDD153 pKa = 3.63LKK155 pKa = 11.19KK156 pKa = 10.38MGEE159 pKa = 4.21GDD161 pKa = 3.83PLVCQKK167 pKa = 10.62GMAVLLEE174 pKa = 4.28ALSVLRR180 pKa = 11.84RR181 pKa = 11.84DD182 pKa = 3.72WGVHH186 pKa = 4.73GQPLLCVVSTTGISEE201 pKa = 4.17TRR203 pKa = 11.84DD204 pKa = 3.16IPLLFYY210 pKa = 10.44PLYY213 pKa = 9.04HH214 pKa = 6.62TLLAVPHH221 pKa = 6.75RR222 pKa = 11.84DD223 pKa = 3.27KK224 pKa = 11.39KK225 pKa = 10.85VMEE228 pKa = 4.48GLLVASGEE236 pKa = 4.17RR237 pKa = 11.84FVLVRR242 pKa = 11.84PSLLWDD248 pKa = 3.51GEE250 pKa = 4.15RR251 pKa = 11.84RR252 pKa = 11.84KK253 pKa = 10.86GKK255 pKa = 10.51GKK257 pKa = 9.96GNGVVRR263 pKa = 11.84VGIADD268 pKa = 3.27ARR270 pKa = 11.84TGVVEE275 pKa = 4.28RR276 pKa = 11.84KK277 pKa = 9.18EE278 pKa = 3.82VGYY281 pKa = 8.78TISRR285 pKa = 11.84EE286 pKa = 4.13AVGGWMFEE294 pKa = 3.89HH295 pKa = 7.31LILRR299 pKa = 11.84AGEE302 pKa = 3.95KK303 pKa = 10.76GLVFEE308 pKa = 5.05EE309 pKa = 4.73KK310 pKa = 10.4AVSLTWW316 pKa = 3.56
Molecular weight: 35.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
453076 |
70 |
6946 |
641.8 |
70.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.096 ± 0.079 | 1.396 ± 0.028 |
5.406 ± 0.046 | 5.64 ± 0.059 |
3.925 ± 0.042 | 7.189 ± 0.063 |
2.555 ± 0.028 | 5.417 ± 0.055 |
3.673 ± 0.046 | 9.866 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.072 ± 0.028 | 3.353 ± 0.035 |
5.507 ± 0.062 | 3.834 ± 0.043 |
5.881 ± 0.053 | 7.884 ± 0.08 |
6.073 ± 0.047 | 6.771 ± 0.054 |
1.593 ± 0.026 | 2.869 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
