
Nitrospirae bacterium HCH-1
Taxonomy: cellular organisms; Bacteria; Nitrospirae; unclassified Nitrospirae
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
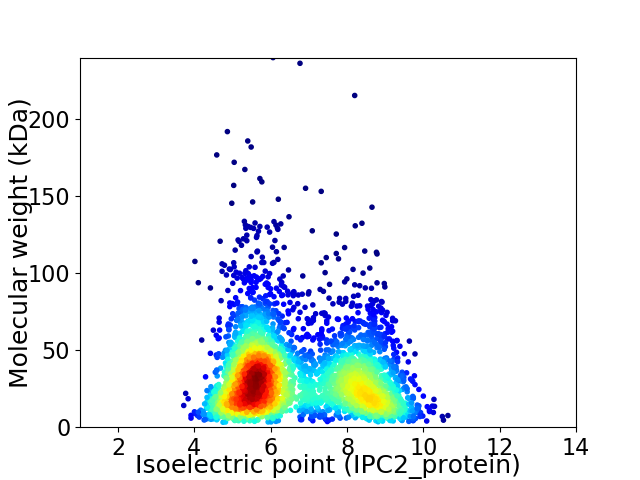
Virtual 2D-PAGE plot for 3334 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A109BTY7|A0A109BTY7_9BACT Potassium transporter TrkA OS=Nitrospirae bacterium HCH-1 OX=1748249 GN=ASN18_3310 PE=4 SV=1
MM1 pKa = 7.03VHH3 pKa = 6.71VDD5 pKa = 5.04DD6 pKa = 5.83PPLDD10 pKa = 3.74TLVGVQLIVPPAPADD25 pKa = 3.54ADD27 pKa = 3.59TAYY30 pKa = 9.75WFNVKK35 pKa = 10.1LAVTEE40 pKa = 3.98QLLVTLVTVYY50 pKa = 9.71VFPDD54 pKa = 3.87NVPPPQVSLTDD65 pKa = 3.08AVYY68 pKa = 9.54PVFGVTVNVVEE79 pKa = 4.71PPLATVFEE87 pKa = 5.01AGDD90 pKa = 3.84MVPPEE95 pKa = 4.6PLTEE99 pKa = 3.96AVTVSAGVNVAAKK112 pKa = 9.83LHH114 pKa = 6.26APVIGAVVNVLPFSVPEE131 pKa = 3.8QPVTVFIWYY140 pKa = 7.69PAFGVSVKK148 pKa = 10.58LAVLPCFTVWLAGLTLPPPPLTDD171 pKa = 3.82AVILL175 pKa = 4.07
MM1 pKa = 7.03VHH3 pKa = 6.71VDD5 pKa = 5.04DD6 pKa = 5.83PPLDD10 pKa = 3.74TLVGVQLIVPPAPADD25 pKa = 3.54ADD27 pKa = 3.59TAYY30 pKa = 9.75WFNVKK35 pKa = 10.1LAVTEE40 pKa = 3.98QLLVTLVTVYY50 pKa = 9.71VFPDD54 pKa = 3.87NVPPPQVSLTDD65 pKa = 3.08AVYY68 pKa = 9.54PVFGVTVNVVEE79 pKa = 4.71PPLATVFEE87 pKa = 5.01AGDD90 pKa = 3.84MVPPEE95 pKa = 4.6PLTEE99 pKa = 3.96AVTVSAGVNVAAKK112 pKa = 9.83LHH114 pKa = 6.26APVIGAVVNVLPFSVPEE131 pKa = 3.8QPVTVFIWYY140 pKa = 7.69PAFGVSVKK148 pKa = 10.58LAVLPCFTVWLAGLTLPPPPLTDD171 pKa = 3.82AVILL175 pKa = 4.07
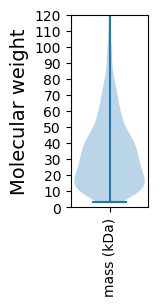
Molecular weight: 18.47 kDa
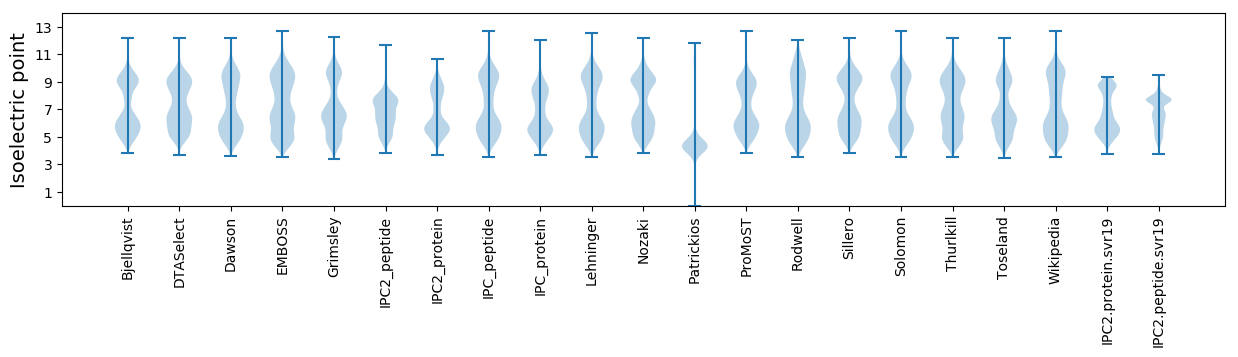
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A109C2R1|A0A109C2R1_9BACT Desulforedoxin OS=Nitrospirae bacterium HCH-1 OX=1748249 GN=ASN18_2726 PE=4 SV=1
MM1 pKa = 7.33AAKK4 pKa = 9.87EE5 pKa = 4.31SKK7 pKa = 10.12EE8 pKa = 3.49EE9 pKa = 3.77HH10 pKa = 5.67RR11 pKa = 11.84RR12 pKa = 11.84LRR14 pKa = 11.84RR15 pKa = 11.84AEE17 pKa = 3.88RR18 pKa = 11.84LLEE21 pKa = 4.16NLSMPSRR28 pKa = 11.84PDD30 pKa = 3.19VLLDD34 pKa = 3.82AVRR37 pKa = 11.84TQAGFAPDD45 pKa = 4.26PLAVTAVILKK55 pKa = 10.72DD56 pKa = 3.51MALSAAVLHH65 pKa = 6.61AANTQLNGWKK75 pKa = 9.74RR76 pKa = 11.84KK77 pKa = 9.53LISIEE82 pKa = 3.87GAVALLGLEE91 pKa = 3.9RR92 pKa = 11.84VRR94 pKa = 11.84AIVAEE99 pKa = 4.16QFLSATLVSQEE110 pKa = 4.73GPLQMVRR117 pKa = 11.84LRR119 pKa = 11.84GVEE122 pKa = 3.87AGRR125 pKa = 11.84VAARR129 pKa = 11.84LARR132 pKa = 11.84EE133 pKa = 4.11LPKK136 pKa = 10.52AAPHH140 pKa = 6.33CLNGYY145 pKa = 8.59LPPVAPDD152 pKa = 3.06EE153 pKa = 4.79AYY155 pKa = 10.37AAGLLHH161 pKa = 7.47DD162 pKa = 5.15CGLVAMMRR170 pKa = 11.84GFPDD174 pKa = 3.35YY175 pKa = 11.41VGFCQEE181 pKa = 3.38MHH183 pKa = 6.21ARR185 pKa = 11.84GGGRR189 pKa = 11.84RR190 pKa = 11.84GGGGEE195 pKa = 3.8RR196 pKa = 11.84AFRR199 pKa = 11.84DD200 pKa = 3.61QPLSGGFFDD209 pKa = 4.29GAEE212 pKa = 4.09MASAGTFLSDD222 pKa = 3.56HH223 pKa = 6.04SHH225 pKa = 6.89PSSDD229 pKa = 3.92GRR231 pKa = 11.84LQPSRR236 pKa = 11.84TEE238 pKa = 3.99GAGTRR243 pKa = 11.84LYY245 pKa = 11.23GSAGHH250 pKa = 5.95TAPFRR255 pKa = 11.84LGLRR259 pKa = 11.84WGVGGGVAGRR269 pKa = 11.84GRR271 pKa = 11.84AHH273 pKa = 7.22HH274 pKa = 6.34LFFWFGSRR282 pKa = 11.84ATGTIARR289 pKa = 11.84PLRR292 pKa = 11.84WGVRR296 pKa = 11.84RR297 pKa = 11.84RR298 pKa = 11.84GWLFSEE304 pKa = 4.99KK305 pKa = 10.27YY306 pKa = 9.22VSIWEE311 pKa = 4.04MKK313 pKa = 9.12RR314 pKa = 11.84AGRR317 pKa = 11.84IHH319 pKa = 7.37RR320 pKa = 11.84GTFLGGG326 pKa = 3.07
MM1 pKa = 7.33AAKK4 pKa = 9.87EE5 pKa = 4.31SKK7 pKa = 10.12EE8 pKa = 3.49EE9 pKa = 3.77HH10 pKa = 5.67RR11 pKa = 11.84RR12 pKa = 11.84LRR14 pKa = 11.84RR15 pKa = 11.84AEE17 pKa = 3.88RR18 pKa = 11.84LLEE21 pKa = 4.16NLSMPSRR28 pKa = 11.84PDD30 pKa = 3.19VLLDD34 pKa = 3.82AVRR37 pKa = 11.84TQAGFAPDD45 pKa = 4.26PLAVTAVILKK55 pKa = 10.72DD56 pKa = 3.51MALSAAVLHH65 pKa = 6.61AANTQLNGWKK75 pKa = 9.74RR76 pKa = 11.84KK77 pKa = 9.53LISIEE82 pKa = 3.87GAVALLGLEE91 pKa = 3.9RR92 pKa = 11.84VRR94 pKa = 11.84AIVAEE99 pKa = 4.16QFLSATLVSQEE110 pKa = 4.73GPLQMVRR117 pKa = 11.84LRR119 pKa = 11.84GVEE122 pKa = 3.87AGRR125 pKa = 11.84VAARR129 pKa = 11.84LARR132 pKa = 11.84EE133 pKa = 4.11LPKK136 pKa = 10.52AAPHH140 pKa = 6.33CLNGYY145 pKa = 8.59LPPVAPDD152 pKa = 3.06EE153 pKa = 4.79AYY155 pKa = 10.37AAGLLHH161 pKa = 7.47DD162 pKa = 5.15CGLVAMMRR170 pKa = 11.84GFPDD174 pKa = 3.35YY175 pKa = 11.41VGFCQEE181 pKa = 3.38MHH183 pKa = 6.21ARR185 pKa = 11.84GGGRR189 pKa = 11.84RR190 pKa = 11.84GGGGEE195 pKa = 3.8RR196 pKa = 11.84AFRR199 pKa = 11.84DD200 pKa = 3.61QPLSGGFFDD209 pKa = 4.29GAEE212 pKa = 4.09MASAGTFLSDD222 pKa = 3.56HH223 pKa = 6.04SHH225 pKa = 6.89PSSDD229 pKa = 3.92GRR231 pKa = 11.84LQPSRR236 pKa = 11.84TEE238 pKa = 3.99GAGTRR243 pKa = 11.84LYY245 pKa = 11.23GSAGHH250 pKa = 5.95TAPFRR255 pKa = 11.84LGLRR259 pKa = 11.84WGVGGGVAGRR269 pKa = 11.84GRR271 pKa = 11.84AHH273 pKa = 7.22HH274 pKa = 6.34LFFWFGSRR282 pKa = 11.84ATGTIARR289 pKa = 11.84PLRR292 pKa = 11.84WGVRR296 pKa = 11.84RR297 pKa = 11.84RR298 pKa = 11.84GWLFSEE304 pKa = 4.99KK305 pKa = 10.27YY306 pKa = 9.22VSIWEE311 pKa = 4.04MKK313 pKa = 9.12RR314 pKa = 11.84AGRR317 pKa = 11.84IHH319 pKa = 7.37RR320 pKa = 11.84GTFLGGG326 pKa = 3.07
Molecular weight: 35.31 kDa
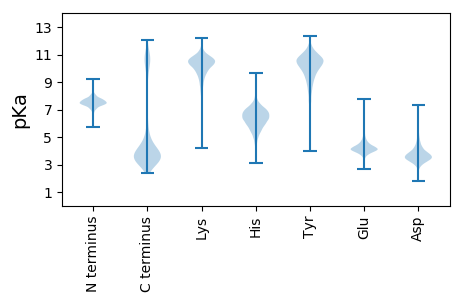
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1080144 |
29 |
2156 |
324.0 |
36.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.903 ± 0.051 | 1.149 ± 0.018 |
5.558 ± 0.03 | 6.239 ± 0.049 |
4.258 ± 0.031 | 6.917 ± 0.039 |
1.889 ± 0.02 | 8.094 ± 0.042 |
6.83 ± 0.038 | 9.335 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.751 ± 0.018 | 4.225 ± 0.028 |
3.811 ± 0.028 | 2.756 ± 0.021 |
4.659 ± 0.034 | 6.414 ± 0.041 |
5.794 ± 0.037 | 6.829 ± 0.031 |
0.941 ± 0.016 | 3.648 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
