
Pontibacter akesuensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Hymenobacteraceae; Pontibacter
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
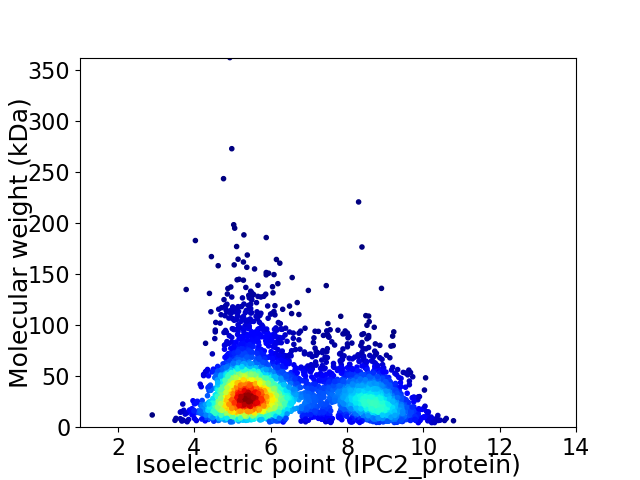
Virtual 2D-PAGE plot for 4073 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I7GRK5|A0A1I7GRK5_9BACT Peptidase family M28 OS=Pontibacter akesuensis OX=388950 GN=SAMN04487941_1211 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.45KK3 pKa = 10.22KK4 pKa = 10.45LLPLRR9 pKa = 11.84LLSFLVCLFFSTAAMAQQVFINEE32 pKa = 3.48IHH34 pKa = 6.59YY35 pKa = 11.24DD36 pKa = 3.57NDD38 pKa = 3.81GTDD41 pKa = 2.88SGEE44 pKa = 4.16AVEE47 pKa = 4.79IAAPSGTDD55 pKa = 3.22LTGWSLVLYY64 pKa = 10.07NGSNNEE70 pKa = 4.15TYY72 pKa = 8.73QTIPLTGAVPASSISYY88 pKa = 10.41GFLSVNTPGIQNGSPDD104 pKa = 4.34AIALVNAAGEE114 pKa = 4.57VVQFLSYY121 pKa = 10.77EE122 pKa = 4.24GTLTAVNGPASGMTSEE138 pKa = 5.46DD139 pKa = 2.92IGVSEE144 pKa = 5.44SSTTPAGYY152 pKa = 10.3SLQLGNIDD160 pKa = 5.23DD161 pKa = 4.88GNASADD167 pKa = 3.92FTWQEE172 pKa = 3.84AATSTFGEE180 pKa = 4.27INNNQVFPLGGGTPDD195 pKa = 4.44PEE197 pKa = 4.59PEE199 pKa = 4.43PEE201 pKa = 4.58PEE203 pKa = 4.27PEE205 pKa = 3.8PAITLVFINEE215 pKa = 3.83LHH217 pKa = 6.51YY218 pKa = 11.53DD219 pKa = 3.63NASTDD224 pKa = 3.17TGEE227 pKa = 4.29GVEE230 pKa = 4.21IAGLAGTNLEE240 pKa = 3.97GWQLVGYY247 pKa = 9.3NGNGGANYY255 pKa = 8.11STTSLSGVIPNQQSGFGTVFFPIPGLQNGSPDD287 pKa = 3.94GIALVNSEE295 pKa = 4.07GQVVQFLSYY304 pKa = 10.08EE305 pKa = 4.09GAFAAVGGPADD316 pKa = 3.5GMMSEE321 pKa = 5.15DD322 pKa = 3.37IGVAEE327 pKa = 5.36AGDD330 pKa = 4.13SPTGYY335 pKa = 10.55SLQLTGSGSSYY346 pKa = 11.5DD347 pKa = 3.55AFTWAASSTATYY359 pKa = 11.06DD360 pKa = 3.43EE361 pKa = 4.69VNNGQTFLPLQDD373 pKa = 3.22VVFINEE379 pKa = 3.68LHH381 pKa = 6.3YY382 pKa = 11.54DD383 pKa = 3.53NDD385 pKa = 4.08GADD388 pKa = 3.08AGEE391 pKa = 4.28GVEE394 pKa = 4.1IAGNAGADD402 pKa = 3.47LSGWQLVLYY411 pKa = 9.68NGNGGTAYY419 pKa = 10.55SALEE423 pKa = 3.98LSGTIPNQDD432 pKa = 2.95NGYY435 pKa = 8.12GTLFFAITGLQNGAPDD451 pKa = 4.36GVALINPDD459 pKa = 3.53GEE461 pKa = 4.64VVQFLSYY468 pKa = 10.34EE469 pKa = 4.24GSFTATDD476 pKa = 3.59GLAAGVTSEE485 pKa = 5.23DD486 pKa = 3.1IGVMEE491 pKa = 5.31SSSTPLNYY499 pKa = 10.22SLQLTGTGTSYY510 pKa = 12.05ADD512 pKa = 3.83FTWGGPITSTYY523 pKa = 11.21NLINTGQSFGGSTQEE538 pKa = 4.4PEE540 pKa = 3.96PEE542 pKa = 4.16VPVEE546 pKa = 3.85GSIAEE551 pKa = 4.11ARR553 pKa = 11.84TLPVGTVVTVSGVLTATNQFGGPAYY578 pKa = 10.5LQDD581 pKa = 3.62ATAGIAVFDD590 pKa = 4.36PQVHH594 pKa = 6.46GDD596 pKa = 3.4GAFAIGDD603 pKa = 4.36SIQITASVEE612 pKa = 4.03LFNQMPEE619 pKa = 4.15LVNITSIEE627 pKa = 4.22SFGPATTTLAPITTTIADD645 pKa = 4.05LNQYY649 pKa = 10.6EE650 pKa = 4.54GMLVTIPGISFTDD663 pKa = 3.71PEE665 pKa = 4.56GLLFPDD671 pKa = 3.73SNYY674 pKa = 10.57EE675 pKa = 3.89ITDD678 pKa = 3.42GTGTVALRR686 pKa = 11.84IDD688 pKa = 4.18GDD690 pKa = 3.85VEE692 pKa = 4.42DD693 pKa = 4.23LVGRR697 pKa = 11.84LKK699 pKa = 10.52PQEE702 pKa = 4.22PVTITGIVSSFRR714 pKa = 11.84GSLQLLPRR722 pKa = 11.84FMADD726 pKa = 3.15LPGTEE731 pKa = 5.23AYY733 pKa = 9.81VQAGSDD739 pKa = 3.32IPVAQTLDD747 pKa = 3.52VMSWNMEE754 pKa = 4.11FFGATQKK761 pKa = 11.25DD762 pKa = 3.76FGPNNEE768 pKa = 4.43LLQLQNAAEE777 pKa = 4.34LLDD780 pKa = 4.48SIRR783 pKa = 11.84ADD785 pKa = 3.36IIAVQEE791 pKa = 4.23VSDD794 pKa = 4.06EE795 pKa = 4.2NQLQQLVDD803 pKa = 3.81MLPGYY808 pKa = 10.88ARR810 pKa = 11.84ICSDD814 pKa = 3.47RR815 pKa = 11.84YY816 pKa = 9.57SYY818 pKa = 11.4SFNGEE823 pKa = 4.01DD824 pKa = 4.38PNFPAQKK831 pKa = 10.47LCFIYY836 pKa = 9.03NTEE839 pKa = 3.94VVTVVDD845 pKa = 3.72EE846 pKa = 4.39RR847 pKa = 11.84VLFEE851 pKa = 4.36EE852 pKa = 5.7LYY854 pKa = 10.31DD855 pKa = 3.74AARR858 pKa = 11.84AGQTDD863 pKa = 4.37LLNNYY868 pKa = 8.99PSGEE872 pKa = 3.94ASSFWSSGRR881 pKa = 11.84LPYY884 pKa = 9.82MVTVDD889 pKa = 3.28ATINGVTEE897 pKa = 5.21RR898 pKa = 11.84IQLINIHH905 pKa = 6.35AKK907 pKa = 10.3SGSNNNDD914 pKa = 3.44LARR917 pKa = 11.84RR918 pKa = 11.84AYY920 pKa = 9.96DD921 pKa = 3.47VQVLKK926 pKa = 11.01DD927 pKa = 3.7SLDD930 pKa = 3.35THH932 pKa = 6.26YY933 pKa = 11.83ANANLILLGDD943 pKa = 3.98YY944 pKa = 11.03NDD946 pKa = 5.54DD947 pKa = 3.25VDD949 pKa = 4.67ASIGSGEE956 pKa = 4.14TVYY959 pKa = 11.02QLFVQDD965 pKa = 2.78IDD967 pKa = 4.39YY968 pKa = 11.35DD969 pKa = 4.23VVTLSLSEE977 pKa = 4.23AGLRR981 pKa = 11.84SYY983 pKa = 8.94ITQEE987 pKa = 3.66NVIDD991 pKa = 5.23HH992 pKa = 5.93ITISDD997 pKa = 3.81EE998 pKa = 4.15LYY1000 pKa = 9.89EE1001 pKa = 4.16EE1002 pKa = 4.67YY1003 pKa = 10.34IEE1005 pKa = 4.61GSEE1008 pKa = 3.92RR1009 pKa = 11.84LIIPFSYY1016 pKa = 10.21IEE1018 pKa = 4.26NYY1020 pKa = 10.85GNTTSDD1026 pKa = 3.52HH1027 pKa = 6.59LPVLVRR1033 pKa = 11.84FEE1035 pKa = 4.23LTAPAPLVVDD1045 pKa = 4.44AGTNQTVYY1053 pKa = 10.5FGYY1056 pKa = 10.65APQACATLTASTPTDD1071 pKa = 3.3GSGMYY1076 pKa = 9.78TYY1078 pKa = 9.35TWSNGQTGQSIQVCPDD1094 pKa = 2.71EE1095 pKa = 4.5TTTYY1099 pKa = 9.72TVTVTDD1105 pKa = 3.67SNGRR1109 pKa = 11.84TGTDD1113 pKa = 3.12EE1114 pKa = 4.29VQVCVVNVVCTKK1126 pKa = 9.96NARR1129 pKa = 11.84TPMVEE1134 pKa = 3.42ICFRR1138 pKa = 11.84PPGRR1142 pKa = 11.84SMEE1145 pKa = 4.31SARR1148 pKa = 11.84TLCVPVQAVEE1158 pKa = 4.18PMLKK1162 pKa = 10.43LGATLGSCGTVACGEE1177 pKa = 4.35EE1178 pKa = 4.4AVEE1181 pKa = 4.74PAPSMPFSITVYY1193 pKa = 10.38PNPLKK1198 pKa = 10.92DD1199 pKa = 3.45FAEE1202 pKa = 4.35VKK1204 pKa = 10.3IEE1206 pKa = 3.8GMVNGDD1212 pKa = 3.88VQVVLYY1218 pKa = 10.26DD1219 pKa = 3.5HH1220 pKa = 7.23FSNEE1224 pKa = 4.01VLSKK1228 pKa = 10.79QISTKK1233 pKa = 10.73DD1234 pKa = 3.36GSFKK1238 pKa = 11.02LDD1240 pKa = 3.6LNSNRR1245 pKa = 11.84LKK1247 pKa = 11.03AGLYY1251 pKa = 7.24YY1252 pKa = 10.8LKK1254 pKa = 10.7VISQNNSQTVRR1265 pKa = 11.84IIKK1268 pKa = 9.55QQ1269 pKa = 2.98
MM1 pKa = 7.47KK2 pKa = 10.45KK3 pKa = 10.22KK4 pKa = 10.45LLPLRR9 pKa = 11.84LLSFLVCLFFSTAAMAQQVFINEE32 pKa = 3.48IHH34 pKa = 6.59YY35 pKa = 11.24DD36 pKa = 3.57NDD38 pKa = 3.81GTDD41 pKa = 2.88SGEE44 pKa = 4.16AVEE47 pKa = 4.79IAAPSGTDD55 pKa = 3.22LTGWSLVLYY64 pKa = 10.07NGSNNEE70 pKa = 4.15TYY72 pKa = 8.73QTIPLTGAVPASSISYY88 pKa = 10.41GFLSVNTPGIQNGSPDD104 pKa = 4.34AIALVNAAGEE114 pKa = 4.57VVQFLSYY121 pKa = 10.77EE122 pKa = 4.24GTLTAVNGPASGMTSEE138 pKa = 5.46DD139 pKa = 2.92IGVSEE144 pKa = 5.44SSTTPAGYY152 pKa = 10.3SLQLGNIDD160 pKa = 5.23DD161 pKa = 4.88GNASADD167 pKa = 3.92FTWQEE172 pKa = 3.84AATSTFGEE180 pKa = 4.27INNNQVFPLGGGTPDD195 pKa = 4.44PEE197 pKa = 4.59PEE199 pKa = 4.43PEE201 pKa = 4.58PEE203 pKa = 4.27PEE205 pKa = 3.8PAITLVFINEE215 pKa = 3.83LHH217 pKa = 6.51YY218 pKa = 11.53DD219 pKa = 3.63NASTDD224 pKa = 3.17TGEE227 pKa = 4.29GVEE230 pKa = 4.21IAGLAGTNLEE240 pKa = 3.97GWQLVGYY247 pKa = 9.3NGNGGANYY255 pKa = 8.11STTSLSGVIPNQQSGFGTVFFPIPGLQNGSPDD287 pKa = 3.94GIALVNSEE295 pKa = 4.07GQVVQFLSYY304 pKa = 10.08EE305 pKa = 4.09GAFAAVGGPADD316 pKa = 3.5GMMSEE321 pKa = 5.15DD322 pKa = 3.37IGVAEE327 pKa = 5.36AGDD330 pKa = 4.13SPTGYY335 pKa = 10.55SLQLTGSGSSYY346 pKa = 11.5DD347 pKa = 3.55AFTWAASSTATYY359 pKa = 11.06DD360 pKa = 3.43EE361 pKa = 4.69VNNGQTFLPLQDD373 pKa = 3.22VVFINEE379 pKa = 3.68LHH381 pKa = 6.3YY382 pKa = 11.54DD383 pKa = 3.53NDD385 pKa = 4.08GADD388 pKa = 3.08AGEE391 pKa = 4.28GVEE394 pKa = 4.1IAGNAGADD402 pKa = 3.47LSGWQLVLYY411 pKa = 9.68NGNGGTAYY419 pKa = 10.55SALEE423 pKa = 3.98LSGTIPNQDD432 pKa = 2.95NGYY435 pKa = 8.12GTLFFAITGLQNGAPDD451 pKa = 4.36GVALINPDD459 pKa = 3.53GEE461 pKa = 4.64VVQFLSYY468 pKa = 10.34EE469 pKa = 4.24GSFTATDD476 pKa = 3.59GLAAGVTSEE485 pKa = 5.23DD486 pKa = 3.1IGVMEE491 pKa = 5.31SSSTPLNYY499 pKa = 10.22SLQLTGTGTSYY510 pKa = 12.05ADD512 pKa = 3.83FTWGGPITSTYY523 pKa = 11.21NLINTGQSFGGSTQEE538 pKa = 4.4PEE540 pKa = 3.96PEE542 pKa = 4.16VPVEE546 pKa = 3.85GSIAEE551 pKa = 4.11ARR553 pKa = 11.84TLPVGTVVTVSGVLTATNQFGGPAYY578 pKa = 10.5LQDD581 pKa = 3.62ATAGIAVFDD590 pKa = 4.36PQVHH594 pKa = 6.46GDD596 pKa = 3.4GAFAIGDD603 pKa = 4.36SIQITASVEE612 pKa = 4.03LFNQMPEE619 pKa = 4.15LVNITSIEE627 pKa = 4.22SFGPATTTLAPITTTIADD645 pKa = 4.05LNQYY649 pKa = 10.6EE650 pKa = 4.54GMLVTIPGISFTDD663 pKa = 3.71PEE665 pKa = 4.56GLLFPDD671 pKa = 3.73SNYY674 pKa = 10.57EE675 pKa = 3.89ITDD678 pKa = 3.42GTGTVALRR686 pKa = 11.84IDD688 pKa = 4.18GDD690 pKa = 3.85VEE692 pKa = 4.42DD693 pKa = 4.23LVGRR697 pKa = 11.84LKK699 pKa = 10.52PQEE702 pKa = 4.22PVTITGIVSSFRR714 pKa = 11.84GSLQLLPRR722 pKa = 11.84FMADD726 pKa = 3.15LPGTEE731 pKa = 5.23AYY733 pKa = 9.81VQAGSDD739 pKa = 3.32IPVAQTLDD747 pKa = 3.52VMSWNMEE754 pKa = 4.11FFGATQKK761 pKa = 11.25DD762 pKa = 3.76FGPNNEE768 pKa = 4.43LLQLQNAAEE777 pKa = 4.34LLDD780 pKa = 4.48SIRR783 pKa = 11.84ADD785 pKa = 3.36IIAVQEE791 pKa = 4.23VSDD794 pKa = 4.06EE795 pKa = 4.2NQLQQLVDD803 pKa = 3.81MLPGYY808 pKa = 10.88ARR810 pKa = 11.84ICSDD814 pKa = 3.47RR815 pKa = 11.84YY816 pKa = 9.57SYY818 pKa = 11.4SFNGEE823 pKa = 4.01DD824 pKa = 4.38PNFPAQKK831 pKa = 10.47LCFIYY836 pKa = 9.03NTEE839 pKa = 3.94VVTVVDD845 pKa = 3.72EE846 pKa = 4.39RR847 pKa = 11.84VLFEE851 pKa = 4.36EE852 pKa = 5.7LYY854 pKa = 10.31DD855 pKa = 3.74AARR858 pKa = 11.84AGQTDD863 pKa = 4.37LLNNYY868 pKa = 8.99PSGEE872 pKa = 3.94ASSFWSSGRR881 pKa = 11.84LPYY884 pKa = 9.82MVTVDD889 pKa = 3.28ATINGVTEE897 pKa = 5.21RR898 pKa = 11.84IQLINIHH905 pKa = 6.35AKK907 pKa = 10.3SGSNNNDD914 pKa = 3.44LARR917 pKa = 11.84RR918 pKa = 11.84AYY920 pKa = 9.96DD921 pKa = 3.47VQVLKK926 pKa = 11.01DD927 pKa = 3.7SLDD930 pKa = 3.35THH932 pKa = 6.26YY933 pKa = 11.83ANANLILLGDD943 pKa = 3.98YY944 pKa = 11.03NDD946 pKa = 5.54DD947 pKa = 3.25VDD949 pKa = 4.67ASIGSGEE956 pKa = 4.14TVYY959 pKa = 11.02QLFVQDD965 pKa = 2.78IDD967 pKa = 4.39YY968 pKa = 11.35DD969 pKa = 4.23VVTLSLSEE977 pKa = 4.23AGLRR981 pKa = 11.84SYY983 pKa = 8.94ITQEE987 pKa = 3.66NVIDD991 pKa = 5.23HH992 pKa = 5.93ITISDD997 pKa = 3.81EE998 pKa = 4.15LYY1000 pKa = 9.89EE1001 pKa = 4.16EE1002 pKa = 4.67YY1003 pKa = 10.34IEE1005 pKa = 4.61GSEE1008 pKa = 3.92RR1009 pKa = 11.84LIIPFSYY1016 pKa = 10.21IEE1018 pKa = 4.26NYY1020 pKa = 10.85GNTTSDD1026 pKa = 3.52HH1027 pKa = 6.59LPVLVRR1033 pKa = 11.84FEE1035 pKa = 4.23LTAPAPLVVDD1045 pKa = 4.44AGTNQTVYY1053 pKa = 10.5FGYY1056 pKa = 10.65APQACATLTASTPTDD1071 pKa = 3.3GSGMYY1076 pKa = 9.78TYY1078 pKa = 9.35TWSNGQTGQSIQVCPDD1094 pKa = 2.71EE1095 pKa = 4.5TTTYY1099 pKa = 9.72TVTVTDD1105 pKa = 3.67SNGRR1109 pKa = 11.84TGTDD1113 pKa = 3.12EE1114 pKa = 4.29VQVCVVNVVCTKK1126 pKa = 9.96NARR1129 pKa = 11.84TPMVEE1134 pKa = 3.42ICFRR1138 pKa = 11.84PPGRR1142 pKa = 11.84SMEE1145 pKa = 4.31SARR1148 pKa = 11.84TLCVPVQAVEE1158 pKa = 4.18PMLKK1162 pKa = 10.43LGATLGSCGTVACGEE1177 pKa = 4.35EE1178 pKa = 4.4AVEE1181 pKa = 4.74PAPSMPFSITVYY1193 pKa = 10.38PNPLKK1198 pKa = 10.92DD1199 pKa = 3.45FAEE1202 pKa = 4.35VKK1204 pKa = 10.3IEE1206 pKa = 3.8GMVNGDD1212 pKa = 3.88VQVVLYY1218 pKa = 10.26DD1219 pKa = 3.5HH1220 pKa = 7.23FSNEE1224 pKa = 4.01VLSKK1228 pKa = 10.79QISTKK1233 pKa = 10.73DD1234 pKa = 3.36GSFKK1238 pKa = 11.02LDD1240 pKa = 3.6LNSNRR1245 pKa = 11.84LKK1247 pKa = 11.03AGLYY1251 pKa = 7.24YY1252 pKa = 10.8LKK1254 pKa = 10.7VISQNNSQTVRR1265 pKa = 11.84IIKK1268 pKa = 9.55QQ1269 pKa = 2.98
Molecular weight: 134.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I7J0S6|A0A1I7J0S6_9BACT Quinol:cytochrome c oxidoreductase monoheme cytochrome subunit OS=Pontibacter akesuensis OX=388950 GN=SAMN04487941_2382 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.54RR3 pKa = 11.84TFQPSQRR10 pKa = 11.84KK11 pKa = 8.72RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.56HH16 pKa = 3.97GFRR19 pKa = 11.84SRR21 pKa = 11.84MEE23 pKa = 3.95TANGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.54GRR39 pKa = 11.84KK40 pKa = 8.64RR41 pKa = 11.84LTVSSEE47 pKa = 3.88KK48 pKa = 9.99RR49 pKa = 11.84HH50 pKa = 5.45KK51 pKa = 10.57AAA53 pKa = 5.14
MM1 pKa = 7.45KK2 pKa = 9.54RR3 pKa = 11.84TFQPSQRR10 pKa = 11.84KK11 pKa = 8.72RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.56HH16 pKa = 3.97GFRR19 pKa = 11.84SRR21 pKa = 11.84MEE23 pKa = 3.95TANGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.54GRR39 pKa = 11.84KK40 pKa = 8.64RR41 pKa = 11.84LTVSSEE47 pKa = 3.88KK48 pKa = 9.99RR49 pKa = 11.84HH50 pKa = 5.45KK51 pKa = 10.57AAA53 pKa = 5.14
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1372960 |
38 |
3596 |
337.1 |
37.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.392 ± 0.04 | 0.712 ± 0.013 |
5.047 ± 0.027 | 6.512 ± 0.047 |
4.483 ± 0.028 | 6.989 ± 0.043 |
2.0 ± 0.022 | 5.894 ± 0.034 |
5.838 ± 0.042 | 10.165 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.41 ± 0.019 | 4.681 ± 0.029 |
4.137 ± 0.025 | 4.383 ± 0.03 |
4.6 ± 0.029 | 5.947 ± 0.035 |
5.674 ± 0.048 | 7.038 ± 0.031 |
1.138 ± 0.017 | 3.961 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
