
Trichoderma asperellum CBS 433.97
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Hypocreaceae; Trichoderma; Trichoderma asperellum
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
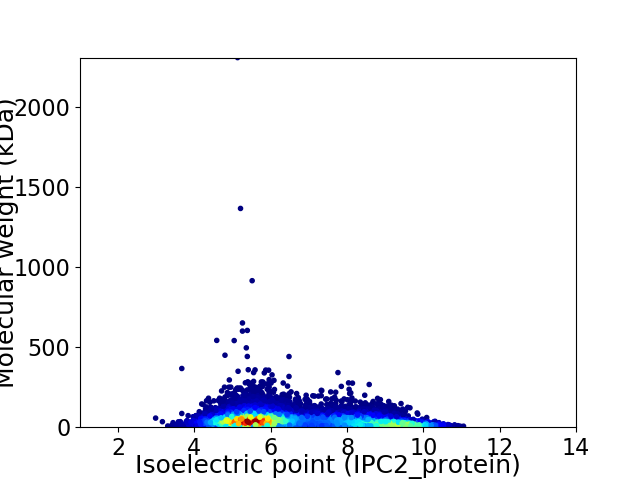
Virtual 2D-PAGE plot for 12547 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T3Z282|A0A2T3Z282_9HYPO Uncharacterized protein OS=Trichoderma asperellum CBS 433.97 OX=1042311 GN=M441DRAFT_60133 PE=4 SV=1
MM1 pKa = 7.61GNYY4 pKa = 9.76SLYY7 pKa = 10.56GFRR10 pKa = 11.84FEE12 pKa = 5.53SEE14 pKa = 4.2SLLLVSDD21 pKa = 3.94QEE23 pKa = 4.32EE24 pKa = 4.44LGNTEE29 pKa = 4.36AEE31 pKa = 4.21YY32 pKa = 10.52EE33 pKa = 4.01QWQNLLEE40 pKa = 4.87LDD42 pKa = 3.76AQASISLQSPNNRR55 pKa = 11.84YY56 pKa = 7.88EE57 pKa = 4.09ANQIPAAEE65 pKa = 4.95HH66 pKa = 6.38ISVQNYY72 pKa = 6.45HH73 pKa = 5.63TQGSDD78 pKa = 2.87NQQTRR83 pKa = 11.84RR84 pKa = 11.84INVAHH89 pKa = 6.75RR90 pKa = 11.84HH91 pKa = 4.17SHH93 pKa = 6.43LPQLIDD99 pKa = 3.08WGSDD103 pKa = 2.9SGSGIEE109 pKa = 5.1DD110 pKa = 2.84IVGIYY115 pKa = 10.83GNDD118 pKa = 3.57PQGLLDD124 pKa = 5.62SPDD127 pKa = 3.42EE128 pKa = 4.83PIWQILYY135 pKa = 10.49SFSYY139 pKa = 10.19QNRR142 pKa = 11.84LDD144 pKa = 4.02PGRR147 pKa = 11.84DD148 pKa = 3.44AYY150 pKa = 10.38EE151 pKa = 4.06HH152 pKa = 5.86TVSAEE157 pKa = 4.0NEE159 pKa = 3.84AGAPHH164 pKa = 6.36ATIKK168 pKa = 11.03NEE170 pKa = 4.25SEE172 pKa = 3.91DD173 pKa = 3.41EE174 pKa = 4.19NYY176 pKa = 10.14PFEE179 pKa = 5.25IMVEE183 pKa = 4.01SGEE186 pKa = 4.21EE187 pKa = 3.9HH188 pKa = 6.69SHH190 pKa = 7.85DD191 pKa = 3.88EE192 pKa = 4.16DD193 pKa = 5.06SIFDD197 pKa = 3.71VVSEE201 pKa = 4.46EE202 pKa = 4.14EE203 pKa = 4.35STDD206 pKa = 3.69DD207 pKa = 3.39EE208 pKa = 5.46CGFEE212 pKa = 5.53SDD214 pKa = 6.1DD215 pKa = 3.84EE216 pKa = 4.82DD217 pKa = 5.71ADD219 pKa = 4.1EE220 pKa = 6.33DD221 pKa = 4.37SDD223 pKa = 5.21SDD225 pKa = 3.61MDD227 pKa = 4.1LSEE230 pKa = 5.92DD231 pKa = 4.31DD232 pKa = 3.71GHH234 pKa = 8.84LEE236 pKa = 3.78LAEE239 pKa = 4.15EE240 pKa = 4.46AGEE243 pKa = 4.24TTYY246 pKa = 11.02MLGGDD251 pKa = 4.07LLVRR255 pKa = 11.84IPTHH259 pKa = 5.84SVFDD263 pKa = 4.06RR264 pKa = 11.84YY265 pKa = 10.91SPEE268 pKa = 3.92EE269 pKa = 3.97YY270 pKa = 10.37LGEE273 pKa = 4.4TILDD277 pKa = 3.82IEE279 pKa = 4.59WPNSGWQNAFNQTEE293 pKa = 4.2QNN295 pKa = 3.41
MM1 pKa = 7.61GNYY4 pKa = 9.76SLYY7 pKa = 10.56GFRR10 pKa = 11.84FEE12 pKa = 5.53SEE14 pKa = 4.2SLLLVSDD21 pKa = 3.94QEE23 pKa = 4.32EE24 pKa = 4.44LGNTEE29 pKa = 4.36AEE31 pKa = 4.21YY32 pKa = 10.52EE33 pKa = 4.01QWQNLLEE40 pKa = 4.87LDD42 pKa = 3.76AQASISLQSPNNRR55 pKa = 11.84YY56 pKa = 7.88EE57 pKa = 4.09ANQIPAAEE65 pKa = 4.95HH66 pKa = 6.38ISVQNYY72 pKa = 6.45HH73 pKa = 5.63TQGSDD78 pKa = 2.87NQQTRR83 pKa = 11.84RR84 pKa = 11.84INVAHH89 pKa = 6.75RR90 pKa = 11.84HH91 pKa = 4.17SHH93 pKa = 6.43LPQLIDD99 pKa = 3.08WGSDD103 pKa = 2.9SGSGIEE109 pKa = 5.1DD110 pKa = 2.84IVGIYY115 pKa = 10.83GNDD118 pKa = 3.57PQGLLDD124 pKa = 5.62SPDD127 pKa = 3.42EE128 pKa = 4.83PIWQILYY135 pKa = 10.49SFSYY139 pKa = 10.19QNRR142 pKa = 11.84LDD144 pKa = 4.02PGRR147 pKa = 11.84DD148 pKa = 3.44AYY150 pKa = 10.38EE151 pKa = 4.06HH152 pKa = 5.86TVSAEE157 pKa = 4.0NEE159 pKa = 3.84AGAPHH164 pKa = 6.36ATIKK168 pKa = 11.03NEE170 pKa = 4.25SEE172 pKa = 3.91DD173 pKa = 3.41EE174 pKa = 4.19NYY176 pKa = 10.14PFEE179 pKa = 5.25IMVEE183 pKa = 4.01SGEE186 pKa = 4.21EE187 pKa = 3.9HH188 pKa = 6.69SHH190 pKa = 7.85DD191 pKa = 3.88EE192 pKa = 4.16DD193 pKa = 5.06SIFDD197 pKa = 3.71VVSEE201 pKa = 4.46EE202 pKa = 4.14EE203 pKa = 4.35STDD206 pKa = 3.69DD207 pKa = 3.39EE208 pKa = 5.46CGFEE212 pKa = 5.53SDD214 pKa = 6.1DD215 pKa = 3.84EE216 pKa = 4.82DD217 pKa = 5.71ADD219 pKa = 4.1EE220 pKa = 6.33DD221 pKa = 4.37SDD223 pKa = 5.21SDD225 pKa = 3.61MDD227 pKa = 4.1LSEE230 pKa = 5.92DD231 pKa = 4.31DD232 pKa = 3.71GHH234 pKa = 8.84LEE236 pKa = 3.78LAEE239 pKa = 4.15EE240 pKa = 4.46AGEE243 pKa = 4.24TTYY246 pKa = 11.02MLGGDD251 pKa = 4.07LLVRR255 pKa = 11.84IPTHH259 pKa = 5.84SVFDD263 pKa = 4.06RR264 pKa = 11.84YY265 pKa = 10.91SPEE268 pKa = 3.92EE269 pKa = 3.97YY270 pKa = 10.37LGEE273 pKa = 4.4TILDD277 pKa = 3.82IEE279 pKa = 4.59WPNSGWQNAFNQTEE293 pKa = 4.2QNN295 pKa = 3.41
Molecular weight: 33.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T3ZA42|A0A2T3ZA42_9HYPO Uncharacterized protein OS=Trichoderma asperellum CBS 433.97 OX=1042311 GN=M441DRAFT_402414 PE=4 SV=1
MM1 pKa = 7.94VSINQLLIGIFFFFFKK17 pKa = 10.94KK18 pKa = 10.39NMQSQTRR25 pKa = 11.84NALFRR30 pKa = 11.84PRR32 pKa = 11.84SSNKK36 pKa = 8.32PVRR39 pKa = 11.84PRR41 pKa = 11.84LHH43 pKa = 6.47KK44 pKa = 10.03VWTVSAISLRR54 pKa = 3.81
MM1 pKa = 7.94VSINQLLIGIFFFFFKK17 pKa = 10.94KK18 pKa = 10.39NMQSQTRR25 pKa = 11.84NALFRR30 pKa = 11.84PRR32 pKa = 11.84SSNKK36 pKa = 8.32PVRR39 pKa = 11.84PRR41 pKa = 11.84LHH43 pKa = 6.47KK44 pKa = 10.03VWTVSAISLRR54 pKa = 3.81
Molecular weight: 6.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5543688 |
49 |
20866 |
441.8 |
48.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.746 ± 0.02 | 1.333 ± 0.01 |
5.635 ± 0.015 | 6.044 ± 0.024 |
3.804 ± 0.014 | 6.751 ± 0.024 |
2.367 ± 0.01 | 5.224 ± 0.019 |
4.905 ± 0.017 | 8.981 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.213 ± 0.009 | 3.777 ± 0.012 |
5.849 ± 0.028 | 4.041 ± 0.017 |
5.947 ± 0.021 | 8.355 ± 0.026 |
5.764 ± 0.016 | 5.973 ± 0.018 |
1.488 ± 0.008 | 2.802 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
