
Raspberry leaf mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Closterovirus
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
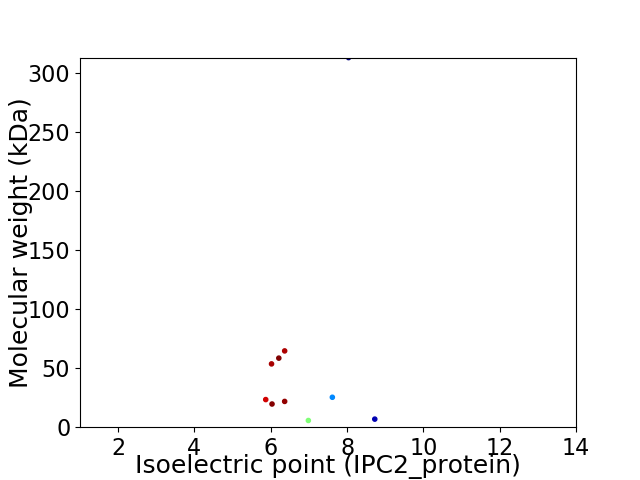
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0MBW5|A0MBW5_9CLOS p7 OS=Raspberry leaf mottle virus OX=326941 PE=4 SV=1
SS1 pKa = 6.97YY2 pKa = 10.13IRR4 pKa = 11.84SQAIPPRR11 pKa = 11.84RR12 pKa = 11.84PSLQEE17 pKa = 3.47NLLSYY22 pKa = 10.25EE23 pKa = 3.78SRR25 pKa = 11.84NFNFIKK31 pKa = 10.44CEE33 pKa = 3.82RR34 pKa = 11.84FSSPKK39 pKa = 10.53LFGAAMASNLLLKK52 pKa = 10.91LFDD55 pKa = 4.0AEE57 pKa = 4.17KK58 pKa = 10.19LAEE61 pKa = 4.11VRR63 pKa = 11.84QSVISISEE71 pKa = 3.91ANIAKK76 pKa = 9.48WLLKK80 pKa = 10.13RR81 pKa = 11.84DD82 pKa = 3.61ASQVKK87 pKa = 10.48ALMTDD92 pKa = 3.55LDD94 pKa = 4.26RR95 pKa = 11.84DD96 pKa = 3.8FDD98 pKa = 4.02IMDD101 pKa = 5.19DD102 pKa = 3.28ISRR105 pKa = 11.84FKK107 pKa = 11.5LMVKK111 pKa = 9.85RR112 pKa = 11.84DD113 pKa = 3.9AKK115 pKa = 11.12VKK117 pKa = 10.8LDD119 pKa = 3.84DD120 pKa = 4.14SCLSKK125 pKa = 10.87HH126 pKa = 6.29PPAQNIMFHH135 pKa = 6.67RR136 pKa = 11.84KK137 pKa = 9.01ALNAVYY143 pKa = 10.1SPCFDD148 pKa = 3.32EE149 pKa = 4.99FKK151 pKa = 11.34NRR153 pKa = 11.84FLYY156 pKa = 10.05CLPPNIVFFTEE167 pKa = 3.89MTNEE171 pKa = 3.86DD172 pKa = 3.45LAEE175 pKa = 4.68IIRR178 pKa = 11.84RR179 pKa = 11.84RR180 pKa = 11.84LGDD183 pKa = 3.71DD184 pKa = 4.04DD185 pKa = 4.41IYY187 pKa = 11.5NIGEE191 pKa = 3.91VDD193 pKa = 4.24FSKK196 pKa = 10.63FDD198 pKa = 3.25KK199 pKa = 11.09SQDD202 pKa = 3.58VFIKK206 pKa = 9.85EE207 pKa = 4.09YY208 pKa = 10.88EE209 pKa = 3.99RR210 pKa = 11.84ALYY213 pKa = 9.66EE214 pKa = 3.96AFGFDD219 pKa = 4.45VEE221 pKa = 4.36LLEE224 pKa = 4.01MWMEE228 pKa = 4.34GEE230 pKa = 4.24YY231 pKa = 10.61NAYY234 pKa = 10.27ASTMDD239 pKa = 3.27SQLSFRR245 pKa = 11.84IEE247 pKa = 3.71NQRR250 pKa = 11.84RR251 pKa = 11.84SGGSNTWIGNSLVTLGLLSMYY272 pKa = 10.64YY273 pKa = 10.2DD274 pKa = 3.38VSKK277 pKa = 10.82FRR279 pKa = 11.84LLLISGDD286 pKa = 3.76DD287 pKa = 3.44SLIYY291 pKa = 10.35SDD293 pKa = 5.55EE294 pKa = 4.36KK295 pKa = 11.09IKK297 pKa = 10.98DD298 pKa = 3.57HH299 pKa = 7.06SSQICLEE306 pKa = 4.29TGFEE310 pKa = 4.48TKK312 pKa = 10.05FLSPSVPYY320 pKa = 9.82FCSKK324 pKa = 10.42FVVQTGSMTYY334 pKa = 9.62FVPDD338 pKa = 4.12PYY340 pKa = 11.41KK341 pKa = 10.99LLVKK345 pKa = 10.63LGGSTPFVTDD355 pKa = 2.64VDD357 pKa = 4.06LFEE360 pKa = 5.86AFVSFRR366 pKa = 11.84DD367 pKa = 3.37LTRR370 pKa = 11.84AFDD373 pKa = 3.67HH374 pKa = 6.56QVMLEE379 pKa = 4.02RR380 pKa = 11.84LCGLVHH386 pKa = 6.61TKK388 pKa = 10.69YY389 pKa = 10.61CFTSGSTLPALCAIHH404 pKa = 7.2CIRR407 pKa = 11.84ANFSSFKK414 pKa = 10.31KK415 pKa = 10.42LYY417 pKa = 9.11PKK419 pKa = 9.68TVGWWLVTSSRR430 pKa = 11.84LKK432 pKa = 10.67FLSKK436 pKa = 10.83LPGLIVSKK444 pKa = 10.67AFSSTGEE451 pKa = 4.15SNYY454 pKa = 9.89FCYY457 pKa = 10.61LRR459 pKa = 11.84DD460 pKa = 3.78SFADD464 pKa = 3.69DD465 pKa = 3.9PGG467 pKa = 4.76
SS1 pKa = 6.97YY2 pKa = 10.13IRR4 pKa = 11.84SQAIPPRR11 pKa = 11.84RR12 pKa = 11.84PSLQEE17 pKa = 3.47NLLSYY22 pKa = 10.25EE23 pKa = 3.78SRR25 pKa = 11.84NFNFIKK31 pKa = 10.44CEE33 pKa = 3.82RR34 pKa = 11.84FSSPKK39 pKa = 10.53LFGAAMASNLLLKK52 pKa = 10.91LFDD55 pKa = 4.0AEE57 pKa = 4.17KK58 pKa = 10.19LAEE61 pKa = 4.11VRR63 pKa = 11.84QSVISISEE71 pKa = 3.91ANIAKK76 pKa = 9.48WLLKK80 pKa = 10.13RR81 pKa = 11.84DD82 pKa = 3.61ASQVKK87 pKa = 10.48ALMTDD92 pKa = 3.55LDD94 pKa = 4.26RR95 pKa = 11.84DD96 pKa = 3.8FDD98 pKa = 4.02IMDD101 pKa = 5.19DD102 pKa = 3.28ISRR105 pKa = 11.84FKK107 pKa = 11.5LMVKK111 pKa = 9.85RR112 pKa = 11.84DD113 pKa = 3.9AKK115 pKa = 11.12VKK117 pKa = 10.8LDD119 pKa = 3.84DD120 pKa = 4.14SCLSKK125 pKa = 10.87HH126 pKa = 6.29PPAQNIMFHH135 pKa = 6.67RR136 pKa = 11.84KK137 pKa = 9.01ALNAVYY143 pKa = 10.1SPCFDD148 pKa = 3.32EE149 pKa = 4.99FKK151 pKa = 11.34NRR153 pKa = 11.84FLYY156 pKa = 10.05CLPPNIVFFTEE167 pKa = 3.89MTNEE171 pKa = 3.86DD172 pKa = 3.45LAEE175 pKa = 4.68IIRR178 pKa = 11.84RR179 pKa = 11.84RR180 pKa = 11.84LGDD183 pKa = 3.71DD184 pKa = 4.04DD185 pKa = 4.41IYY187 pKa = 11.5NIGEE191 pKa = 3.91VDD193 pKa = 4.24FSKK196 pKa = 10.63FDD198 pKa = 3.25KK199 pKa = 11.09SQDD202 pKa = 3.58VFIKK206 pKa = 9.85EE207 pKa = 4.09YY208 pKa = 10.88EE209 pKa = 3.99RR210 pKa = 11.84ALYY213 pKa = 9.66EE214 pKa = 3.96AFGFDD219 pKa = 4.45VEE221 pKa = 4.36LLEE224 pKa = 4.01MWMEE228 pKa = 4.34GEE230 pKa = 4.24YY231 pKa = 10.61NAYY234 pKa = 10.27ASTMDD239 pKa = 3.27SQLSFRR245 pKa = 11.84IEE247 pKa = 3.71NQRR250 pKa = 11.84RR251 pKa = 11.84SGGSNTWIGNSLVTLGLLSMYY272 pKa = 10.64YY273 pKa = 10.2DD274 pKa = 3.38VSKK277 pKa = 10.82FRR279 pKa = 11.84LLLISGDD286 pKa = 3.76DD287 pKa = 3.44SLIYY291 pKa = 10.35SDD293 pKa = 5.55EE294 pKa = 4.36KK295 pKa = 11.09IKK297 pKa = 10.98DD298 pKa = 3.57HH299 pKa = 7.06SSQICLEE306 pKa = 4.29TGFEE310 pKa = 4.48TKK312 pKa = 10.05FLSPSVPYY320 pKa = 9.82FCSKK324 pKa = 10.42FVVQTGSMTYY334 pKa = 9.62FVPDD338 pKa = 4.12PYY340 pKa = 11.41KK341 pKa = 10.99LLVKK345 pKa = 10.63LGGSTPFVTDD355 pKa = 2.64VDD357 pKa = 4.06LFEE360 pKa = 5.86AFVSFRR366 pKa = 11.84DD367 pKa = 3.37LTRR370 pKa = 11.84AFDD373 pKa = 3.67HH374 pKa = 6.56QVMLEE379 pKa = 4.02RR380 pKa = 11.84LCGLVHH386 pKa = 6.61TKK388 pKa = 10.69YY389 pKa = 10.61CFTSGSTLPALCAIHH404 pKa = 7.2CIRR407 pKa = 11.84ANFSSFKK414 pKa = 10.31KK415 pKa = 10.42LYY417 pKa = 9.11PKK419 pKa = 9.68TVGWWLVTSSRR430 pKa = 11.84LKK432 pKa = 10.67FLSKK436 pKa = 10.83LPGLIVSKK444 pKa = 10.67AFSSTGEE451 pKa = 4.15SNYY454 pKa = 9.89FCYY457 pKa = 10.61LRR459 pKa = 11.84DD460 pKa = 3.78SFADD464 pKa = 3.69DD465 pKa = 3.9PGG467 pKa = 4.76
Molecular weight: 53.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0MBW6|A0MBW6_9CLOS p6 OS=Raspberry leaf mottle virus OX=326941 PE=4 SV=1
MM1 pKa = 8.18PITEE5 pKa = 4.41SEE7 pKa = 4.75TPFSLAASPSSPPLRR22 pKa = 11.84STRR25 pKa = 11.84VPKK28 pKa = 10.68SSILAFSLLLMLTLCLISAFFISCFRR54 pKa = 11.84FHH56 pKa = 7.27RR57 pKa = 11.84FCRR60 pKa = 11.84LL61 pKa = 2.93
MM1 pKa = 8.18PITEE5 pKa = 4.41SEE7 pKa = 4.75TPFSLAASPSSPPLRR22 pKa = 11.84STRR25 pKa = 11.84VPKK28 pKa = 10.68SSILAFSLLLMLTLCLISAFFISCFRR54 pKa = 11.84FHH56 pKa = 7.27RR57 pKa = 11.84FCRR60 pKa = 11.84LL61 pKa = 2.93
Molecular weight: 6.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5318 |
51 |
2819 |
531.8 |
59.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.826 ± 0.537 | 2.219 ± 0.194 |
5.792 ± 0.288 | 4.494 ± 0.44 |
5.923 ± 0.655 | 5.491 ± 0.376 |
2.144 ± 0.14 | 4.87 ± 0.638 |
5.096 ± 0.406 | 10.361 ± 0.338 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.636 ± 0.417 | 3.291 ± 0.316 |
4.964 ± 0.361 | 1.824 ± 0.155 |
6.074 ± 0.465 | 10.624 ± 0.782 |
5.716 ± 0.62 | 8.274 ± 0.728 |
0.696 ± 0.113 | 3.686 ± 0.29 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
