
Aspergillus brasiliensis (strain CBS 101740 / IMI 381727 / IBT 21946)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus; Aspergillus brasiliensis
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
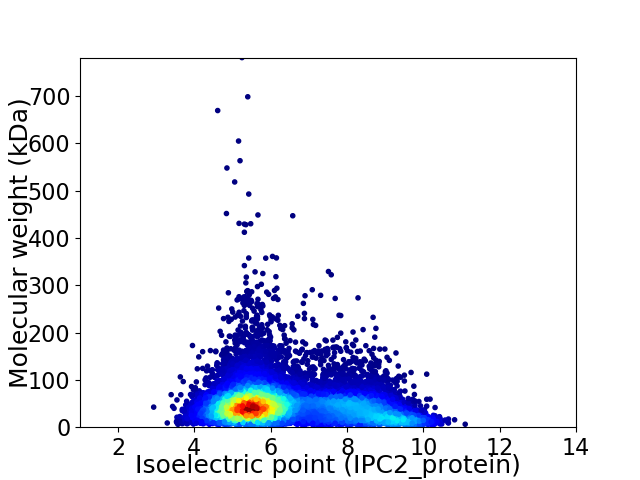
Virtual 2D-PAGE plot for 12982 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L9URS8|A0A1L9URS8_ASPBC Uncharacterized protein OS=Aspergillus brasiliensis (strain CBS 101740 / IMI 381727 / IBT 21946) OX=767769 GN=ASPBRDRAFT_193058 PE=4 SV=1
MM1 pKa = 7.18VSSSVIVLTLWAALVSASPVADD23 pKa = 3.73PLVTPAPKK31 pKa = 10.7LEE33 pKa = 4.33DD34 pKa = 3.46LEE36 pKa = 4.92KK37 pKa = 10.62RR38 pKa = 11.84ATSCTFSGSKK48 pKa = 9.97GASSASKK55 pKa = 10.7SKK57 pKa = 9.53TSCSTIVLSDD67 pKa = 3.44VAVPSGTTLDD77 pKa = 3.9LTDD80 pKa = 5.4LNDD83 pKa = 3.69GTHH86 pKa = 6.48VIFEE90 pKa = 4.49GEE92 pKa = 4.06TTFGYY97 pKa = 10.42KK98 pKa = 9.1EE99 pKa = 3.82WSGPLVSVSGTDD111 pKa = 3.23ITVTGADD118 pKa = 3.38GAYY121 pKa = 10.71LNGDD125 pKa = 3.92GSRR128 pKa = 11.84WWDD131 pKa = 3.38DD132 pKa = 3.26EE133 pKa = 4.13GSNGGKK139 pKa = 8.24TKK141 pKa = 10.59PKK143 pKa = 10.12FFYY146 pKa = 10.74AHH148 pKa = 7.7DD149 pKa = 4.24LTSSTISGIYY159 pKa = 8.66IQNSPVQVFSIDD171 pKa = 3.1GSTDD175 pKa = 3.07LTLEE179 pKa = 5.37DD180 pKa = 3.42ITIDD184 pKa = 3.96NSDD187 pKa = 3.9GDD189 pKa = 4.52DD190 pKa = 3.38EE191 pKa = 5.12GAANTDD197 pKa = 3.17GFDD200 pKa = 3.88IGDD203 pKa = 3.68STYY206 pKa = 9.73ITITGANVYY215 pKa = 10.14NQDD218 pKa = 3.1DD219 pKa = 4.24CVAVNSGEE227 pKa = 4.07NIYY230 pKa = 10.7FSGGVCSGGHH240 pKa = 5.7GLSIGSVGGRR250 pKa = 11.84SDD252 pKa = 3.45NTVKK256 pKa = 10.81NVTFYY261 pKa = 11.3NSEE264 pKa = 3.84IKK266 pKa = 10.65NSQNGVRR273 pKa = 11.84IKK275 pKa = 9.88TIYY278 pKa = 10.53GDD280 pKa = 3.51TGSVSEE286 pKa = 4.22VTYY289 pKa = 11.1KK290 pKa = 10.76EE291 pKa = 3.78ITLSEE296 pKa = 3.79ITKK299 pKa = 10.57YY300 pKa = 10.95GIIVQQNYY308 pKa = 10.46DD309 pKa = 3.91DD310 pKa = 4.27TSEE313 pKa = 4.37SPTDD317 pKa = 4.3GITIEE322 pKa = 5.64DD323 pKa = 3.85FVLDD327 pKa = 3.86NVQGSVEE334 pKa = 4.1SSGTNIYY341 pKa = 9.42IVCGSDD347 pKa = 4.0SCTDD351 pKa = 3.47WTWTDD356 pKa = 2.93VDD358 pKa = 3.88VTGGKK363 pKa = 8.27TSSSCEE369 pKa = 3.93NVPDD373 pKa = 6.1DD374 pKa = 4.49ISCC377 pKa = 3.93
MM1 pKa = 7.18VSSSVIVLTLWAALVSASPVADD23 pKa = 3.73PLVTPAPKK31 pKa = 10.7LEE33 pKa = 4.33DD34 pKa = 3.46LEE36 pKa = 4.92KK37 pKa = 10.62RR38 pKa = 11.84ATSCTFSGSKK48 pKa = 9.97GASSASKK55 pKa = 10.7SKK57 pKa = 9.53TSCSTIVLSDD67 pKa = 3.44VAVPSGTTLDD77 pKa = 3.9LTDD80 pKa = 5.4LNDD83 pKa = 3.69GTHH86 pKa = 6.48VIFEE90 pKa = 4.49GEE92 pKa = 4.06TTFGYY97 pKa = 10.42KK98 pKa = 9.1EE99 pKa = 3.82WSGPLVSVSGTDD111 pKa = 3.23ITVTGADD118 pKa = 3.38GAYY121 pKa = 10.71LNGDD125 pKa = 3.92GSRR128 pKa = 11.84WWDD131 pKa = 3.38DD132 pKa = 3.26EE133 pKa = 4.13GSNGGKK139 pKa = 8.24TKK141 pKa = 10.59PKK143 pKa = 10.12FFYY146 pKa = 10.74AHH148 pKa = 7.7DD149 pKa = 4.24LTSSTISGIYY159 pKa = 8.66IQNSPVQVFSIDD171 pKa = 3.1GSTDD175 pKa = 3.07LTLEE179 pKa = 5.37DD180 pKa = 3.42ITIDD184 pKa = 3.96NSDD187 pKa = 3.9GDD189 pKa = 4.52DD190 pKa = 3.38EE191 pKa = 5.12GAANTDD197 pKa = 3.17GFDD200 pKa = 3.88IGDD203 pKa = 3.68STYY206 pKa = 9.73ITITGANVYY215 pKa = 10.14NQDD218 pKa = 3.1DD219 pKa = 4.24CVAVNSGEE227 pKa = 4.07NIYY230 pKa = 10.7FSGGVCSGGHH240 pKa = 5.7GLSIGSVGGRR250 pKa = 11.84SDD252 pKa = 3.45NTVKK256 pKa = 10.81NVTFYY261 pKa = 11.3NSEE264 pKa = 3.84IKK266 pKa = 10.65NSQNGVRR273 pKa = 11.84IKK275 pKa = 9.88TIYY278 pKa = 10.53GDD280 pKa = 3.51TGSVSEE286 pKa = 4.22VTYY289 pKa = 11.1KK290 pKa = 10.76EE291 pKa = 3.78ITLSEE296 pKa = 3.79ITKK299 pKa = 10.57YY300 pKa = 10.95GIIVQQNYY308 pKa = 10.46DD309 pKa = 3.91DD310 pKa = 4.27TSEE313 pKa = 4.37SPTDD317 pKa = 4.3GITIEE322 pKa = 5.64DD323 pKa = 3.85FVLDD327 pKa = 3.86NVQGSVEE334 pKa = 4.1SSGTNIYY341 pKa = 9.42IVCGSDD347 pKa = 4.0SCTDD351 pKa = 3.47WTWTDD356 pKa = 2.93VDD358 pKa = 3.88VTGGKK363 pKa = 8.27TSSSCEE369 pKa = 3.93NVPDD373 pKa = 6.1DD374 pKa = 4.49ISCC377 pKa = 3.93
Molecular weight: 39.56 kDa
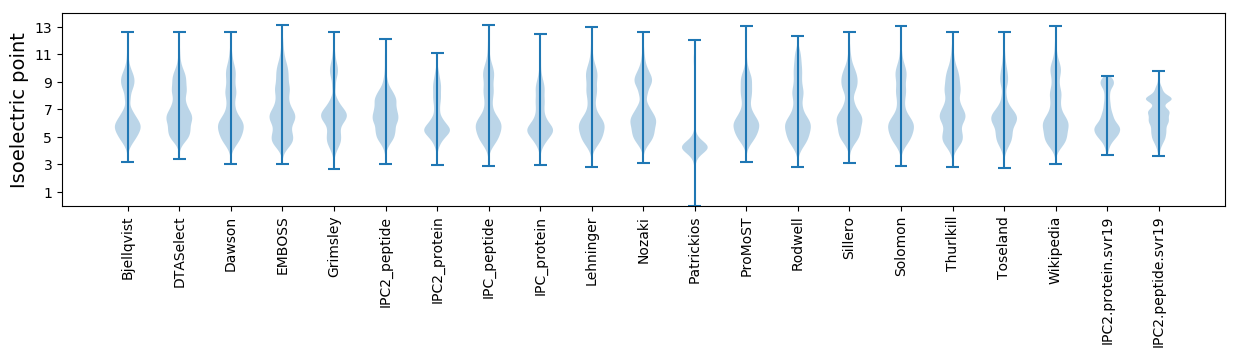
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L9UCD6|A0A1L9UCD6_ASPBC Component of oligomeric Golgi complex 2 OS=Aspergillus brasiliensis (strain CBS 101740 / IMI 381727 / IBT 21946) OX=767769 GN=ASPBRDRAFT_57044 PE=3 SV=1
MM1 pKa = 7.88PSHH4 pKa = 6.91KK5 pKa = 10.39SFRR8 pKa = 11.84TKK10 pKa = 10.45QKK12 pKa = 9.54LAKK15 pKa = 9.74AQRR18 pKa = 11.84QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.78RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.25WRR45 pKa = 11.84KK46 pKa = 9.64SRR48 pKa = 11.84LGVV51 pKa = 3.24
MM1 pKa = 7.88PSHH4 pKa = 6.91KK5 pKa = 10.39SFRR8 pKa = 11.84TKK10 pKa = 10.45QKK12 pKa = 9.54LAKK15 pKa = 9.74AQRR18 pKa = 11.84QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.78RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.25WRR45 pKa = 11.84KK46 pKa = 9.64SRR48 pKa = 11.84LGVV51 pKa = 3.24
Molecular weight: 6.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6024818 |
49 |
7067 |
464.1 |
51.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.422 ± 0.018 | 1.345 ± 0.008 |
5.59 ± 0.013 | 6.04 ± 0.019 |
3.749 ± 0.013 | 6.782 ± 0.02 |
2.49 ± 0.009 | 4.994 ± 0.016 |
4.357 ± 0.017 | 9.205 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.184 ± 0.008 | 3.55 ± 0.011 |
6.024 ± 0.02 | 4.017 ± 0.016 |
6.137 ± 0.018 | 8.316 ± 0.027 |
6.061 ± 0.017 | 6.262 ± 0.016 |
1.526 ± 0.008 | 2.951 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
