
Monosporascus sp. MC13-8B
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Xylariomycetidae; Xylariales; Xylariales incertae sedis; Monosporascus; unclassified Monosporascus
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
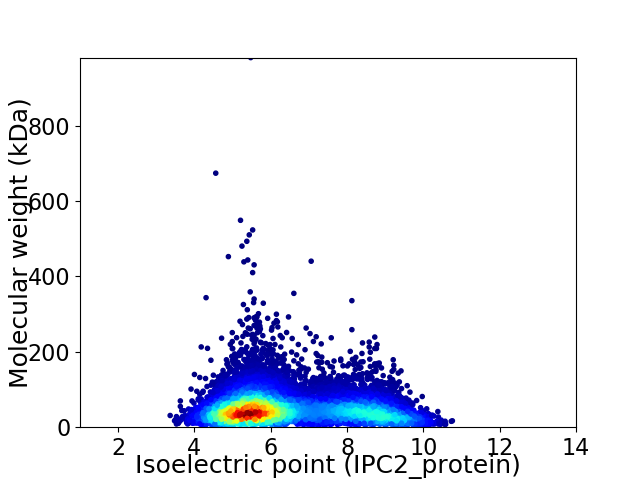
Virtual 2D-PAGE plot for 10637 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q4UQP1|A0A4Q4UQP1_9PEZI Uncharacterized protein OS=Monosporascus sp. MC13-8B OX=2211646 GN=DL766_007648 PE=4 SV=1
MM1 pKa = 7.86HH2 pKa = 7.14ITTILSLTILSALGAAEE19 pKa = 4.45SNVDD23 pKa = 3.59RR24 pKa = 11.84DD25 pKa = 4.46DD26 pKa = 4.05VPSACLASCQSTIDD40 pKa = 3.44LSQRR44 pKa = 11.84CDD46 pKa = 3.21RR47 pKa = 11.84QTDD50 pKa = 3.19DD51 pKa = 4.06DD52 pKa = 4.82NIYY55 pKa = 10.33RR56 pKa = 11.84GCVCGAQDD64 pKa = 4.37AQRR67 pKa = 11.84QMDD70 pKa = 3.35QCAACVRR77 pKa = 11.84VNGMSGPVDD86 pKa = 3.37NDD88 pKa = 3.02VAEE91 pKa = 4.39LMNDD95 pKa = 3.75CGWDD99 pKa = 3.5FNTASGSATAVTSMTSRR116 pKa = 11.84TTAGSTPTPVVSTSTGASTTATITQTPTGASGSGATEE153 pKa = 3.91SSTTNSDD160 pKa = 3.06NAAPAATAGLAGLLVAAGAAVALFF184 pKa = 3.95
MM1 pKa = 7.86HH2 pKa = 7.14ITTILSLTILSALGAAEE19 pKa = 4.45SNVDD23 pKa = 3.59RR24 pKa = 11.84DD25 pKa = 4.46DD26 pKa = 4.05VPSACLASCQSTIDD40 pKa = 3.44LSQRR44 pKa = 11.84CDD46 pKa = 3.21RR47 pKa = 11.84QTDD50 pKa = 3.19DD51 pKa = 4.06DD52 pKa = 4.82NIYY55 pKa = 10.33RR56 pKa = 11.84GCVCGAQDD64 pKa = 4.37AQRR67 pKa = 11.84QMDD70 pKa = 3.35QCAACVRR77 pKa = 11.84VNGMSGPVDD86 pKa = 3.37NDD88 pKa = 3.02VAEE91 pKa = 4.39LMNDD95 pKa = 3.75CGWDD99 pKa = 3.5FNTASGSATAVTSMTSRR116 pKa = 11.84TTAGSTPTPVVSTSTGASTTATITQTPTGASGSGATEE153 pKa = 3.91SSTTNSDD160 pKa = 3.06NAAPAATAGLAGLLVAAGAAVALFF184 pKa = 3.95
Molecular weight: 18.39 kDa
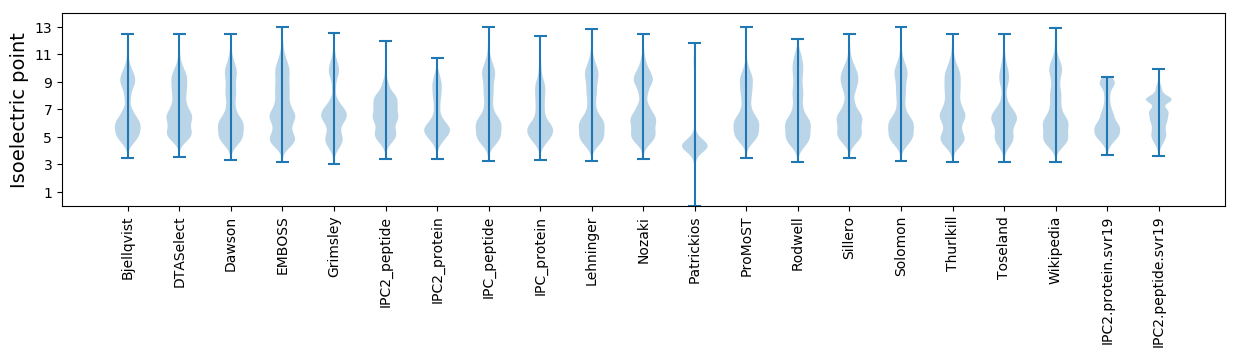
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q4W046|A0A4Q4W046_9PEZI Uncharacterized protein OS=Monosporascus sp. MC13-8B OX=2211646 GN=DL766_000428 PE=4 SV=1
MM1 pKa = 7.68PLCGCKK7 pKa = 9.76LHH9 pKa = 6.47VYY11 pKa = 10.05RR12 pKa = 11.84FRR14 pKa = 11.84LCRR17 pKa = 11.84HH18 pKa = 5.57YY19 pKa = 11.29GNPIKK24 pKa = 10.55HH25 pKa = 5.74EE26 pKa = 4.2CWRR29 pKa = 11.84ADD31 pKa = 3.48LRR33 pKa = 11.84TKK35 pKa = 10.13SPCMGSLFPCGKK47 pKa = 9.0PVKK50 pKa = 9.84AKK52 pKa = 10.22HH53 pKa = 6.26SRR55 pKa = 11.84DD56 pKa = 3.77GLCDD60 pKa = 2.91SCAAYY65 pKa = 9.65FAGFGDD71 pKa = 3.96RR72 pKa = 11.84SNEE75 pKa = 3.62VLAEE79 pKa = 3.77FLEE82 pKa = 4.6YY83 pKa = 10.48KK84 pKa = 10.41EE85 pKa = 3.99SRR87 pKa = 11.84GWRR90 pKa = 11.84KK91 pKa = 9.74KK92 pKa = 10.09RR93 pKa = 11.84VNPRR97 pKa = 11.84QVAPEE102 pKa = 3.97KK103 pKa = 10.33FVRR106 pKa = 11.84PDD108 pKa = 3.12ILLHH112 pKa = 5.78NSSVRR117 pKa = 11.84RR118 pKa = 11.84GGAQPFRR125 pKa = 11.84PQSPMRR131 pKa = 11.84ATLALLPRR139 pKa = 11.84NSHH142 pKa = 6.79LPQPPPPVRR151 pKa = 11.84QTQSGEE157 pKa = 3.49RR158 pKa = 11.84RR159 pKa = 11.84AEE161 pKa = 3.65RR162 pKa = 11.84RR163 pKa = 11.84ARR165 pKa = 11.84TPFEE169 pKa = 3.76HH170 pKa = 7.29ALPGDD175 pKa = 4.06CAASQSSSLILGDD188 pKa = 3.79RR189 pKa = 11.84LLMPNQGVYY198 pKa = 9.36TLPRR202 pKa = 11.84GHH204 pKa = 5.95STEE207 pKa = 4.09PCPLARR213 pKa = 11.84RR214 pKa = 11.84ITEE217 pKa = 3.61EE218 pKa = 4.32AEE220 pKa = 3.88RR221 pKa = 11.84LFPNPEE227 pKa = 3.56SAMYY231 pKa = 9.92PGVPKK236 pKa = 10.78LSAAPKK242 pKa = 8.16KK243 pKa = 9.88HH244 pKa = 5.78RR245 pKa = 11.84RR246 pKa = 11.84NHH248 pKa = 5.44PRR250 pKa = 11.84LPRR253 pKa = 11.84GNDD256 pKa = 2.78NKK258 pKa = 10.81APEE261 pKa = 4.5LTVVPAVIVSGGKK274 pKa = 9.68AFKK277 pKa = 10.4HH278 pKa = 5.78RR279 pKa = 11.84ADD281 pKa = 3.51AATAATRR288 pKa = 11.84SPSEE292 pKa = 4.25PLPAPTPSRR301 pKa = 11.84FFVLASPDD309 pKa = 3.28AMSPTGCSAHH319 pKa = 5.73VRR321 pKa = 11.84SLSLDD326 pKa = 3.25QTVAMPPPQRR336 pKa = 11.84LEE338 pKa = 3.95GVLAASCPQHH348 pKa = 6.55GEE350 pKa = 4.1RR351 pKa = 11.84YY352 pKa = 10.26DD353 pKa = 3.93FGCGEE358 pKa = 4.17CRR360 pKa = 11.84GGLFRR365 pKa = 11.84EE366 pKa = 4.22RR367 pKa = 11.84GLPTEE372 pKa = 4.23EE373 pKa = 3.45HH374 pKa = 6.33RR375 pKa = 11.84EE376 pKa = 4.24RR377 pKa = 11.84YY378 pKa = 9.77RR379 pKa = 11.84SMTPVSVAAAEE390 pKa = 4.21TSNNRR395 pKa = 11.84LGHH398 pKa = 5.91CRR400 pKa = 11.84STPLVVSVTTPRR412 pKa = 11.84RR413 pKa = 11.84SSPVPRR419 pKa = 11.84KK420 pKa = 9.53ACARR424 pKa = 11.84AAPSRR429 pKa = 11.84GTCVSPVTRR438 pKa = 11.84RR439 pKa = 11.84RR440 pKa = 11.84PSRR443 pKa = 11.84NRR445 pKa = 11.84SRR447 pKa = 11.84AVGSEE452 pKa = 4.0DD453 pKa = 3.26
MM1 pKa = 7.68PLCGCKK7 pKa = 9.76LHH9 pKa = 6.47VYY11 pKa = 10.05RR12 pKa = 11.84FRR14 pKa = 11.84LCRR17 pKa = 11.84HH18 pKa = 5.57YY19 pKa = 11.29GNPIKK24 pKa = 10.55HH25 pKa = 5.74EE26 pKa = 4.2CWRR29 pKa = 11.84ADD31 pKa = 3.48LRR33 pKa = 11.84TKK35 pKa = 10.13SPCMGSLFPCGKK47 pKa = 9.0PVKK50 pKa = 9.84AKK52 pKa = 10.22HH53 pKa = 6.26SRR55 pKa = 11.84DD56 pKa = 3.77GLCDD60 pKa = 2.91SCAAYY65 pKa = 9.65FAGFGDD71 pKa = 3.96RR72 pKa = 11.84SNEE75 pKa = 3.62VLAEE79 pKa = 3.77FLEE82 pKa = 4.6YY83 pKa = 10.48KK84 pKa = 10.41EE85 pKa = 3.99SRR87 pKa = 11.84GWRR90 pKa = 11.84KK91 pKa = 9.74KK92 pKa = 10.09RR93 pKa = 11.84VNPRR97 pKa = 11.84QVAPEE102 pKa = 3.97KK103 pKa = 10.33FVRR106 pKa = 11.84PDD108 pKa = 3.12ILLHH112 pKa = 5.78NSSVRR117 pKa = 11.84RR118 pKa = 11.84GGAQPFRR125 pKa = 11.84PQSPMRR131 pKa = 11.84ATLALLPRR139 pKa = 11.84NSHH142 pKa = 6.79LPQPPPPVRR151 pKa = 11.84QTQSGEE157 pKa = 3.49RR158 pKa = 11.84RR159 pKa = 11.84AEE161 pKa = 3.65RR162 pKa = 11.84RR163 pKa = 11.84ARR165 pKa = 11.84TPFEE169 pKa = 3.76HH170 pKa = 7.29ALPGDD175 pKa = 4.06CAASQSSSLILGDD188 pKa = 3.79RR189 pKa = 11.84LLMPNQGVYY198 pKa = 9.36TLPRR202 pKa = 11.84GHH204 pKa = 5.95STEE207 pKa = 4.09PCPLARR213 pKa = 11.84RR214 pKa = 11.84ITEE217 pKa = 3.61EE218 pKa = 4.32AEE220 pKa = 3.88RR221 pKa = 11.84LFPNPEE227 pKa = 3.56SAMYY231 pKa = 9.92PGVPKK236 pKa = 10.78LSAAPKK242 pKa = 8.16KK243 pKa = 9.88HH244 pKa = 5.78RR245 pKa = 11.84RR246 pKa = 11.84NHH248 pKa = 5.44PRR250 pKa = 11.84LPRR253 pKa = 11.84GNDD256 pKa = 2.78NKK258 pKa = 10.81APEE261 pKa = 4.5LTVVPAVIVSGGKK274 pKa = 9.68AFKK277 pKa = 10.4HH278 pKa = 5.78RR279 pKa = 11.84ADD281 pKa = 3.51AATAATRR288 pKa = 11.84SPSEE292 pKa = 4.25PLPAPTPSRR301 pKa = 11.84FFVLASPDD309 pKa = 3.28AMSPTGCSAHH319 pKa = 5.73VRR321 pKa = 11.84SLSLDD326 pKa = 3.25QTVAMPPPQRR336 pKa = 11.84LEE338 pKa = 3.95GVLAASCPQHH348 pKa = 6.55GEE350 pKa = 4.1RR351 pKa = 11.84YY352 pKa = 10.26DD353 pKa = 3.93FGCGEE358 pKa = 4.17CRR360 pKa = 11.84GGLFRR365 pKa = 11.84EE366 pKa = 4.22RR367 pKa = 11.84GLPTEE372 pKa = 4.23EE373 pKa = 3.45HH374 pKa = 6.33RR375 pKa = 11.84EE376 pKa = 4.24RR377 pKa = 11.84YY378 pKa = 9.77RR379 pKa = 11.84SMTPVSVAAAEE390 pKa = 4.21TSNNRR395 pKa = 11.84LGHH398 pKa = 5.91CRR400 pKa = 11.84STPLVVSVTTPRR412 pKa = 11.84RR413 pKa = 11.84SSPVPRR419 pKa = 11.84KK420 pKa = 9.53ACARR424 pKa = 11.84AAPSRR429 pKa = 11.84GTCVSPVTRR438 pKa = 11.84RR439 pKa = 11.84RR440 pKa = 11.84PSRR443 pKa = 11.84NRR445 pKa = 11.84SRR447 pKa = 11.84AVGSEE452 pKa = 4.0DD453 pKa = 3.26
Molecular weight: 49.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5133785 |
66 |
8847 |
482.6 |
53.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.304 ± 0.024 | 1.157 ± 0.008 |
5.896 ± 0.018 | 6.363 ± 0.024 |
3.508 ± 0.016 | 7.385 ± 0.022 |
2.338 ± 0.01 | 4.446 ± 0.015 |
4.601 ± 0.022 | 8.644 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.092 ± 0.008 | 3.49 ± 0.012 |
6.335 ± 0.027 | 3.855 ± 0.016 |
6.696 ± 0.022 | 7.877 ± 0.023 |
5.739 ± 0.017 | 6.151 ± 0.019 |
1.432 ± 0.007 | 2.691 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
