
Torque teno midi virus 15
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus
Average proteome isoelectric point is 7.74
Get precalculated fractions of proteins
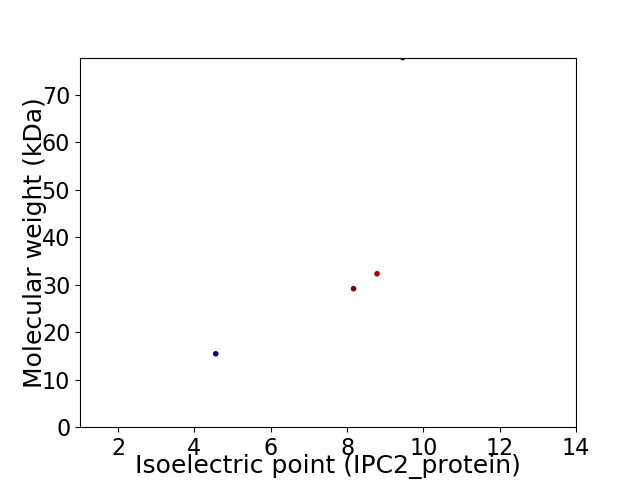
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B6ZK29|B6ZK29_9VIRU Capsid protein OS=Torque teno midi virus 15 OX=2065056 PE=3 SV=1
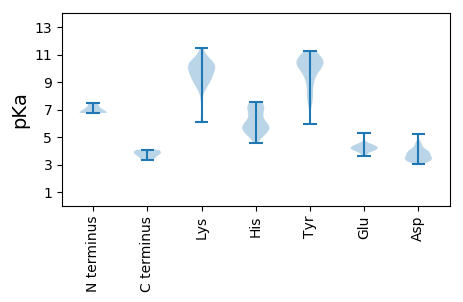
MM1 pKa = 6.75QHH3 pKa = 6.0YY4 pKa = 7.81PPPKK8 pKa = 9.36LYY10 pKa = 10.5KK11 pKa = 10.15IPFLDD16 pKa = 3.2AWGKK20 pKa = 6.09QQQWISSVEE29 pKa = 4.11SQHH32 pKa = 7.15DD33 pKa = 3.83LWCGCHH39 pKa = 5.72HH40 pKa = 7.25WFAHH44 pKa = 5.72FLDD47 pKa = 4.22SVVPLDD53 pKa = 3.3SRR55 pKa = 11.84NRR57 pKa = 11.84GKK59 pKa = 9.17TIQEE63 pKa = 4.05IIEE66 pKa = 4.19QSTHH70 pKa = 5.2QLRR73 pKa = 11.84ALPALPLKK81 pKa = 9.59PWPGDD86 pKa = 3.36TTGDD90 pKa = 3.53EE91 pKa = 4.46KK92 pKa = 11.46DD93 pKa = 3.53PSEE96 pKa = 4.46DD97 pKa = 3.25AGKK100 pKa = 10.53EE101 pKa = 4.05EE102 pKa = 4.27PQNTEE107 pKa = 3.62NQDD110 pKa = 3.37FTDD113 pKa = 4.0VMPEE117 pKa = 3.98DD118 pKa = 3.94EE119 pKa = 4.46LAEE122 pKa = 4.1ILEE125 pKa = 4.32NAARR129 pKa = 11.84DD130 pKa = 3.85EE131 pKa = 4.27NVPGG135 pKa = 3.84
MM1 pKa = 6.75QHH3 pKa = 6.0YY4 pKa = 7.81PPPKK8 pKa = 9.36LYY10 pKa = 10.5KK11 pKa = 10.15IPFLDD16 pKa = 3.2AWGKK20 pKa = 6.09QQQWISSVEE29 pKa = 4.11SQHH32 pKa = 7.15DD33 pKa = 3.83LWCGCHH39 pKa = 5.72HH40 pKa = 7.25WFAHH44 pKa = 5.72FLDD47 pKa = 4.22SVVPLDD53 pKa = 3.3SRR55 pKa = 11.84NRR57 pKa = 11.84GKK59 pKa = 9.17TIQEE63 pKa = 4.05IIEE66 pKa = 4.19QSTHH70 pKa = 5.2QLRR73 pKa = 11.84ALPALPLKK81 pKa = 9.59PWPGDD86 pKa = 3.36TTGDD90 pKa = 3.53EE91 pKa = 4.46KK92 pKa = 11.46DD93 pKa = 3.53PSEE96 pKa = 4.46DD97 pKa = 3.25AGKK100 pKa = 10.53EE101 pKa = 4.05EE102 pKa = 4.27PQNTEE107 pKa = 3.62NQDD110 pKa = 3.37FTDD113 pKa = 4.0VMPEE117 pKa = 3.98DD118 pKa = 3.94EE119 pKa = 4.46LAEE122 pKa = 4.1ILEE125 pKa = 4.32NAARR129 pKa = 11.84DD130 pKa = 3.85EE131 pKa = 4.27NVPGG135 pKa = 3.84
Molecular weight: 15.46 kDa
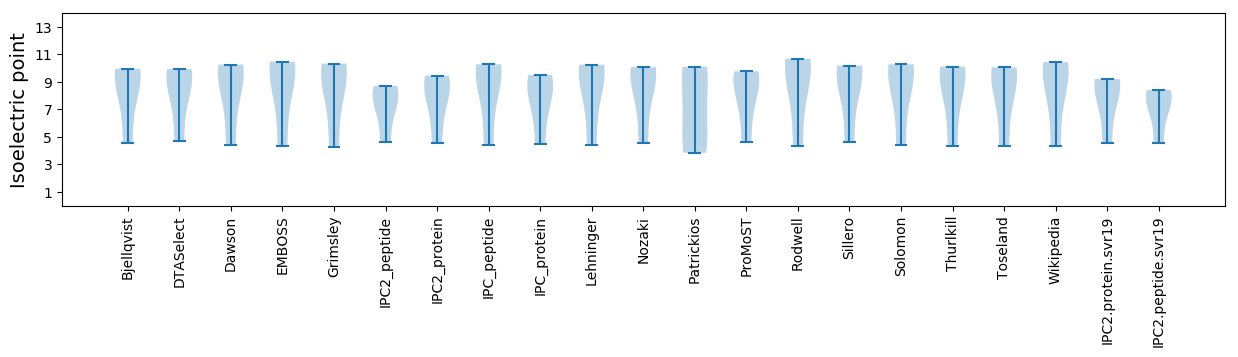
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B6ZK29|B6ZK29_9VIRU Capsid protein OS=Torque teno midi virus 15 OX=2065056 PE=3 SV=1
MM1 pKa = 7.48AWRR4 pKa = 11.84YY5 pKa = 5.97YY6 pKa = 8.56WRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.27RR11 pKa = 11.84PFRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84WKK18 pKa = 10.42RR19 pKa = 11.84RR20 pKa = 11.84ATKK23 pKa = 9.44YY24 pKa = 10.06RR25 pKa = 11.84KK26 pKa = 7.96PRR28 pKa = 11.84LYY30 pKa = 9.92RR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 9.4ARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84TRR39 pKa = 11.84RR40 pKa = 11.84NPRR43 pKa = 11.84KK44 pKa = 9.15RR45 pKa = 11.84RR46 pKa = 11.84QRR48 pKa = 11.84RR49 pKa = 11.84KK50 pKa = 8.74RR51 pKa = 11.84PRR53 pKa = 11.84VRR55 pKa = 11.84RR56 pKa = 11.84KK57 pKa = 9.95RR58 pKa = 11.84QTLPLKK64 pKa = 9.48QWQPEE69 pKa = 4.26SIRR72 pKa = 11.84RR73 pKa = 11.84CKK75 pKa = 10.37IIGHH79 pKa = 5.37TPIILGCSDD88 pKa = 3.91RR89 pKa = 11.84QIHH92 pKa = 6.93SYY94 pKa = 9.45TYY96 pKa = 7.64WQDD99 pKa = 3.53YY100 pKa = 9.34YY101 pKa = 10.67IHH103 pKa = 6.89PRR105 pKa = 11.84CAAGGGFAVMKK116 pKa = 10.21WSLAYY121 pKa = 10.39LYY123 pKa = 10.82EE124 pKa = 4.36DD125 pKa = 4.1FKK127 pKa = 11.11RR128 pKa = 11.84RR129 pKa = 11.84KK130 pKa = 10.02NIWTKK135 pKa = 9.86SNCMYY140 pKa = 10.68DD141 pKa = 3.59LCRR144 pKa = 11.84YY145 pKa = 7.7TGTKK149 pKa = 9.77LKK151 pKa = 10.26LYY153 pKa = 10.15KK154 pKa = 10.32HH155 pKa = 5.67EE156 pKa = 4.17TVDD159 pKa = 4.55FIVSFSRR166 pKa = 11.84NYY168 pKa = 10.42PMVVDD173 pKa = 4.52EE174 pKa = 4.26NTYY177 pKa = 8.91MHH179 pKa = 6.52THH181 pKa = 6.56PSIMLLKK188 pKa = 9.3KK189 pKa = 9.86HH190 pKa = 6.74RR191 pKa = 11.84IIIPSKK197 pKa = 8.67RR198 pKa = 11.84TNPHH202 pKa = 4.57GKK204 pKa = 8.9NYY206 pKa = 10.62KK207 pKa = 9.93KK208 pKa = 9.4ITVRR212 pKa = 11.84PPRR215 pKa = 11.84QLINKK220 pKa = 8.14WFFQKK225 pKa = 10.64NFNDD229 pKa = 3.08QGLFLLTATAADD241 pKa = 3.76LMYY244 pKa = 10.22PHH246 pKa = 7.58LGPYY250 pKa = 9.49SKK252 pKa = 10.81NRR254 pKa = 11.84LVTITILNTQHH265 pKa = 7.55FYY267 pKa = 10.38QQPNWGTATGDD278 pKa = 3.55YY279 pKa = 11.2SFGTTNFSTWGGAKK293 pKa = 9.58TYY295 pKa = 10.38IDD297 pKa = 4.67NKK299 pKa = 9.38LTDD302 pKa = 4.46YY303 pKa = 10.68NPSMDD308 pKa = 4.16MLKK311 pKa = 10.58QNNKK315 pKa = 9.24HH316 pKa = 5.63NASNGIFNKK325 pKa = 9.73EE326 pKa = 3.98FPNIQYY332 pKa = 8.84WKK334 pKa = 8.81TITNPTHH341 pKa = 6.31PTPTMQVQYY350 pKa = 11.2NPARR354 pKa = 11.84DD355 pKa = 3.43TGNGNKK361 pKa = 9.63VYY363 pKa = 10.32FISTLQTSWAPSTTDD378 pKa = 5.25KK379 pKa = 11.43VMTWTGEE386 pKa = 4.14PLWLQLYY393 pKa = 10.32GLTDD397 pKa = 4.0YY398 pKa = 10.79ILKK401 pKa = 10.8VKK403 pKa = 8.84GTSSALQDD411 pKa = 4.53AICVIEE417 pKa = 4.25SPYY420 pKa = 10.31IEE422 pKa = 4.6PQTHH426 pKa = 5.21NKK428 pKa = 9.74KK429 pKa = 10.31FVFVGNPFMNGLTEE443 pKa = 5.21CGNQPPGWLLSKK455 pKa = 10.2WFLTLEE461 pKa = 4.1DD462 pKa = 3.99QMPILNEE469 pKa = 4.08IIKK472 pKa = 10.22SGPYY476 pKa = 9.34VARR479 pKa = 11.84QDD481 pKa = 3.86GPRR484 pKa = 11.84CTWEE488 pKa = 3.6LHH490 pKa = 5.4CDD492 pKa = 3.22YY493 pKa = 11.09RR494 pKa = 11.84SFFKK498 pKa = 10.3WGGAQPPLQNICDD511 pKa = 3.76PHH513 pKa = 7.56LQEE516 pKa = 4.76TFPVPDD522 pKa = 3.79TVKK525 pKa = 9.29QTIQIIDD532 pKa = 3.74PQKK535 pKa = 10.58QIPEE539 pKa = 4.37TLFHH543 pKa = 5.97SWDD546 pKa = 3.44YY547 pKa = 11.26RR548 pKa = 11.84RR549 pKa = 11.84GLITKK554 pKa = 7.58TALKK558 pKa = 10.42RR559 pKa = 11.84MYY561 pKa = 10.8KK562 pKa = 10.22NISDD566 pKa = 4.49ADD568 pKa = 3.78SSSTDD573 pKa = 3.09GSEE576 pKa = 5.06KK577 pKa = 10.62SDD579 pKa = 3.43TPRR582 pKa = 11.84KK583 pKa = 8.77KK584 pKa = 10.15RR585 pKa = 11.84KK586 pKa = 9.9RR587 pKa = 11.84GDD589 pKa = 3.54PPCPEE594 pKa = 4.28EE595 pKa = 4.27KK596 pKa = 10.4DD597 pKa = 3.34NYY599 pKa = 10.0IIQNLQEE606 pKa = 4.12ICQAPQEE613 pKa = 4.33TSGDD617 pKa = 3.83LEE619 pKa = 4.29QQLHH623 pKa = 5.56NLRR626 pKa = 11.84EE627 pKa = 3.92QQLNKK632 pKa = 10.51NISLLKK638 pKa = 10.1LLAMLKK644 pKa = 9.91QKK646 pKa = 9.98QLQMQLAMGMMEE658 pKa = 4.08
MM1 pKa = 7.48AWRR4 pKa = 11.84YY5 pKa = 5.97YY6 pKa = 8.56WRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.27RR11 pKa = 11.84PFRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84WKK18 pKa = 10.42RR19 pKa = 11.84RR20 pKa = 11.84ATKK23 pKa = 9.44YY24 pKa = 10.06RR25 pKa = 11.84KK26 pKa = 7.96PRR28 pKa = 11.84LYY30 pKa = 9.92RR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 9.4ARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84TRR39 pKa = 11.84RR40 pKa = 11.84NPRR43 pKa = 11.84KK44 pKa = 9.15RR45 pKa = 11.84RR46 pKa = 11.84QRR48 pKa = 11.84RR49 pKa = 11.84KK50 pKa = 8.74RR51 pKa = 11.84PRR53 pKa = 11.84VRR55 pKa = 11.84RR56 pKa = 11.84KK57 pKa = 9.95RR58 pKa = 11.84QTLPLKK64 pKa = 9.48QWQPEE69 pKa = 4.26SIRR72 pKa = 11.84RR73 pKa = 11.84CKK75 pKa = 10.37IIGHH79 pKa = 5.37TPIILGCSDD88 pKa = 3.91RR89 pKa = 11.84QIHH92 pKa = 6.93SYY94 pKa = 9.45TYY96 pKa = 7.64WQDD99 pKa = 3.53YY100 pKa = 9.34YY101 pKa = 10.67IHH103 pKa = 6.89PRR105 pKa = 11.84CAAGGGFAVMKK116 pKa = 10.21WSLAYY121 pKa = 10.39LYY123 pKa = 10.82EE124 pKa = 4.36DD125 pKa = 4.1FKK127 pKa = 11.11RR128 pKa = 11.84RR129 pKa = 11.84KK130 pKa = 10.02NIWTKK135 pKa = 9.86SNCMYY140 pKa = 10.68DD141 pKa = 3.59LCRR144 pKa = 11.84YY145 pKa = 7.7TGTKK149 pKa = 9.77LKK151 pKa = 10.26LYY153 pKa = 10.15KK154 pKa = 10.32HH155 pKa = 5.67EE156 pKa = 4.17TVDD159 pKa = 4.55FIVSFSRR166 pKa = 11.84NYY168 pKa = 10.42PMVVDD173 pKa = 4.52EE174 pKa = 4.26NTYY177 pKa = 8.91MHH179 pKa = 6.52THH181 pKa = 6.56PSIMLLKK188 pKa = 9.3KK189 pKa = 9.86HH190 pKa = 6.74RR191 pKa = 11.84IIIPSKK197 pKa = 8.67RR198 pKa = 11.84TNPHH202 pKa = 4.57GKK204 pKa = 8.9NYY206 pKa = 10.62KK207 pKa = 9.93KK208 pKa = 9.4ITVRR212 pKa = 11.84PPRR215 pKa = 11.84QLINKK220 pKa = 8.14WFFQKK225 pKa = 10.64NFNDD229 pKa = 3.08QGLFLLTATAADD241 pKa = 3.76LMYY244 pKa = 10.22PHH246 pKa = 7.58LGPYY250 pKa = 9.49SKK252 pKa = 10.81NRR254 pKa = 11.84LVTITILNTQHH265 pKa = 7.55FYY267 pKa = 10.38QQPNWGTATGDD278 pKa = 3.55YY279 pKa = 11.2SFGTTNFSTWGGAKK293 pKa = 9.58TYY295 pKa = 10.38IDD297 pKa = 4.67NKK299 pKa = 9.38LTDD302 pKa = 4.46YY303 pKa = 10.68NPSMDD308 pKa = 4.16MLKK311 pKa = 10.58QNNKK315 pKa = 9.24HH316 pKa = 5.63NASNGIFNKK325 pKa = 9.73EE326 pKa = 3.98FPNIQYY332 pKa = 8.84WKK334 pKa = 8.81TITNPTHH341 pKa = 6.31PTPTMQVQYY350 pKa = 11.2NPARR354 pKa = 11.84DD355 pKa = 3.43TGNGNKK361 pKa = 9.63VYY363 pKa = 10.32FISTLQTSWAPSTTDD378 pKa = 5.25KK379 pKa = 11.43VMTWTGEE386 pKa = 4.14PLWLQLYY393 pKa = 10.32GLTDD397 pKa = 4.0YY398 pKa = 10.79ILKK401 pKa = 10.8VKK403 pKa = 8.84GTSSALQDD411 pKa = 4.53AICVIEE417 pKa = 4.25SPYY420 pKa = 10.31IEE422 pKa = 4.6PQTHH426 pKa = 5.21NKK428 pKa = 9.74KK429 pKa = 10.31FVFVGNPFMNGLTEE443 pKa = 5.21CGNQPPGWLLSKK455 pKa = 10.2WFLTLEE461 pKa = 4.1DD462 pKa = 3.99QMPILNEE469 pKa = 4.08IIKK472 pKa = 10.22SGPYY476 pKa = 9.34VARR479 pKa = 11.84QDD481 pKa = 3.86GPRR484 pKa = 11.84CTWEE488 pKa = 3.6LHH490 pKa = 5.4CDD492 pKa = 3.22YY493 pKa = 11.09RR494 pKa = 11.84SFFKK498 pKa = 10.3WGGAQPPLQNICDD511 pKa = 3.76PHH513 pKa = 7.56LQEE516 pKa = 4.76TFPVPDD522 pKa = 3.79TVKK525 pKa = 9.29QTIQIIDD532 pKa = 3.74PQKK535 pKa = 10.58QIPEE539 pKa = 4.37TLFHH543 pKa = 5.97SWDD546 pKa = 3.44YY547 pKa = 11.26RR548 pKa = 11.84RR549 pKa = 11.84GLITKK554 pKa = 7.58TALKK558 pKa = 10.42RR559 pKa = 11.84MYY561 pKa = 10.8KK562 pKa = 10.22NISDD566 pKa = 4.49ADD568 pKa = 3.78SSSTDD573 pKa = 3.09GSEE576 pKa = 5.06KK577 pKa = 10.62SDD579 pKa = 3.43TPRR582 pKa = 11.84KK583 pKa = 8.77KK584 pKa = 10.15RR585 pKa = 11.84KK586 pKa = 9.9RR587 pKa = 11.84GDD589 pKa = 3.54PPCPEE594 pKa = 4.28EE595 pKa = 4.27KK596 pKa = 10.4DD597 pKa = 3.34NYY599 pKa = 10.0IIQNLQEE606 pKa = 4.12ICQAPQEE613 pKa = 4.33TSGDD617 pKa = 3.83LEE619 pKa = 4.29QQLHH623 pKa = 5.56NLRR626 pKa = 11.84EE627 pKa = 3.92QQLNKK632 pKa = 10.51NISLLKK638 pKa = 10.1LLAMLKK644 pKa = 9.91QKK646 pKa = 9.98QLQMQLAMGMMEE658 pKa = 4.08
Molecular weight: 77.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1329 |
135 |
658 |
332.3 |
38.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.117 ± 0.87 | 1.505 ± 0.177 |
5.267 ± 0.62 | 5.342 ± 1.113 |
3.085 ± 0.382 | 4.515 ± 0.359 |
3.386 ± 0.441 | 5.267 ± 0.439 |
8.126 ± 0.585 | 7.449 ± 0.498 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.107 ± 0.433 | 4.59 ± 0.635 |
7.976 ± 1.051 | 6.922 ± 0.289 |
6.622 ± 0.837 | 6.02 ± 1.316 |
7.299 ± 0.61 | 3.01 ± 0.338 |
2.784 ± 0.231 | 3.612 ± 0.833 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
