
Hubei sobemo-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.68
Get precalculated fractions of proteins
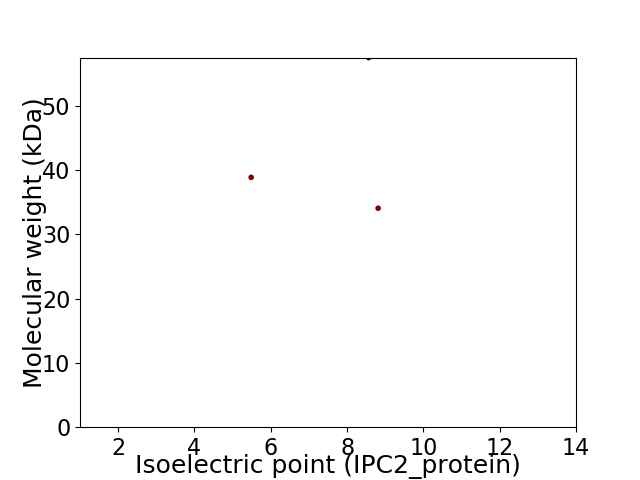
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
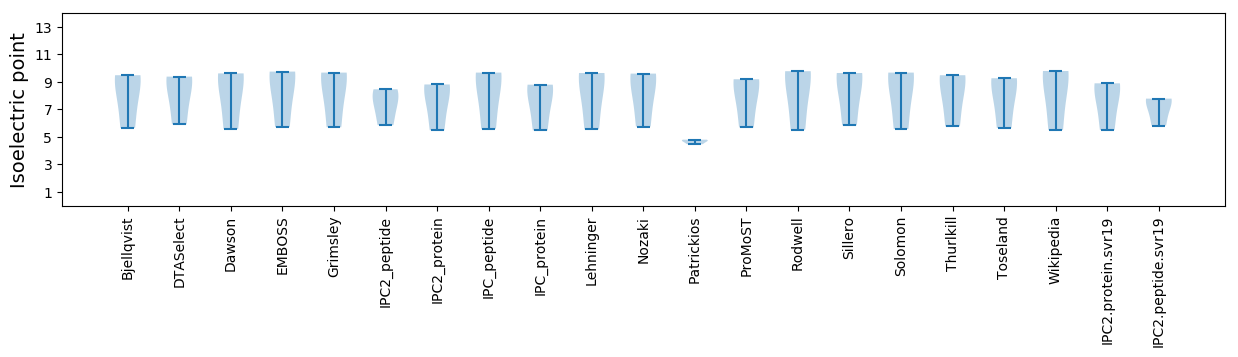
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KER8|A0A1L3KER8_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 1 OX=1923194 PE=4 SV=1
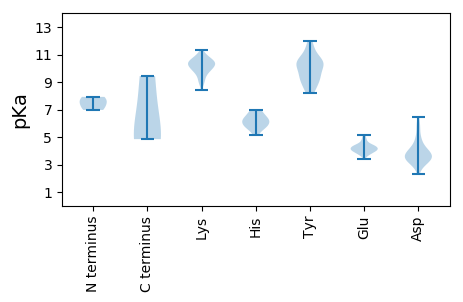
MM1 pKa = 7.92RR2 pKa = 11.84IAATNKK8 pKa = 9.93EE9 pKa = 4.25LLEE12 pKa = 4.11LHH14 pKa = 6.32WSLVEE19 pKa = 3.89NAAVARR25 pKa = 11.84LQLLEE30 pKa = 4.13EE31 pKa = 4.17VDD33 pKa = 4.23YY34 pKa = 11.56EE35 pKa = 4.38DD36 pKa = 4.7LRR38 pKa = 11.84AMSASEE44 pKa = 4.04LVEE47 pKa = 4.22GGYY50 pKa = 9.06TDD52 pKa = 3.46EE53 pKa = 4.25VRR55 pKa = 11.84VFVKK59 pKa = 10.98NEE61 pKa = 3.37LHH63 pKa = 6.44SRR65 pKa = 11.84AKK67 pKa = 9.71VAEE70 pKa = 3.79GRR72 pKa = 11.84MRR74 pKa = 11.84LIMSVSVVDD83 pKa = 4.17QIVEE87 pKa = 4.15RR88 pKa = 11.84VLNSAQNQLEE98 pKa = 3.93ISKK101 pKa = 10.02WEE103 pKa = 4.35TISSKK108 pKa = 10.72PGMGLDD114 pKa = 4.14DD115 pKa = 5.75DD116 pKa = 4.9SLFSIQEE123 pKa = 3.97QVHH126 pKa = 5.91KK127 pKa = 10.43LGSGVGDD134 pKa = 3.82PVSSDD139 pKa = 2.33ISGFDD144 pKa = 3.03WSVPQWALDD153 pKa = 3.27LDD155 pKa = 4.0ARR157 pKa = 11.84VRR159 pKa = 11.84AALSGAPTWLRR170 pKa = 11.84VHH172 pKa = 6.74EE173 pKa = 4.41KK174 pKa = 10.55RR175 pKa = 11.84AVCLGLSQLVFSDD188 pKa = 3.88GVVWDD193 pKa = 3.65QLEE196 pKa = 4.17PGIQKK201 pKa = 10.25SGSYY205 pKa = 8.29NTSSTNSRR213 pKa = 11.84IRR215 pKa = 11.84VMLAWLVARR224 pKa = 11.84RR225 pKa = 11.84IPQSRR230 pKa = 11.84YY231 pKa = 8.66GGRR234 pKa = 11.84DD235 pKa = 3.1ALEE238 pKa = 4.37GGGRR242 pKa = 11.84AMAMGDD248 pKa = 3.63DD249 pKa = 4.64AIEE252 pKa = 4.19TTTCMEE258 pKa = 4.25LLKK261 pKa = 10.69PNYY264 pKa = 9.06EE265 pKa = 3.84RR266 pKa = 11.84LGFVLKK272 pKa = 10.34EE273 pKa = 3.71VSRR276 pKa = 11.84DD277 pKa = 3.3IEE279 pKa = 4.06FCAYY283 pKa = 10.32AFDD286 pKa = 4.48LAQGYY291 pKa = 9.28RR292 pKa = 11.84PLRR295 pKa = 11.84VVKK298 pKa = 9.74MVAGLLRR305 pKa = 11.84TAVRR309 pKa = 11.84DD310 pKa = 3.71EE311 pKa = 3.85QHH313 pKa = 6.42EE314 pKa = 4.38EE315 pKa = 3.9EE316 pKa = 4.7LRR318 pKa = 11.84VALEE322 pKa = 3.79YY323 pKa = 10.63EE324 pKa = 4.38LRR326 pKa = 11.84HH327 pKa = 6.09SDD329 pKa = 3.0KK330 pKa = 10.96QAWAKK335 pKa = 10.26HH336 pKa = 5.15VIRR339 pKa = 11.84ASGWGARR346 pKa = 11.84KK347 pKa = 9.44
MM1 pKa = 7.92RR2 pKa = 11.84IAATNKK8 pKa = 9.93EE9 pKa = 4.25LLEE12 pKa = 4.11LHH14 pKa = 6.32WSLVEE19 pKa = 3.89NAAVARR25 pKa = 11.84LQLLEE30 pKa = 4.13EE31 pKa = 4.17VDD33 pKa = 4.23YY34 pKa = 11.56EE35 pKa = 4.38DD36 pKa = 4.7LRR38 pKa = 11.84AMSASEE44 pKa = 4.04LVEE47 pKa = 4.22GGYY50 pKa = 9.06TDD52 pKa = 3.46EE53 pKa = 4.25VRR55 pKa = 11.84VFVKK59 pKa = 10.98NEE61 pKa = 3.37LHH63 pKa = 6.44SRR65 pKa = 11.84AKK67 pKa = 9.71VAEE70 pKa = 3.79GRR72 pKa = 11.84MRR74 pKa = 11.84LIMSVSVVDD83 pKa = 4.17QIVEE87 pKa = 4.15RR88 pKa = 11.84VLNSAQNQLEE98 pKa = 3.93ISKK101 pKa = 10.02WEE103 pKa = 4.35TISSKK108 pKa = 10.72PGMGLDD114 pKa = 4.14DD115 pKa = 5.75DD116 pKa = 4.9SLFSIQEE123 pKa = 3.97QVHH126 pKa = 5.91KK127 pKa = 10.43LGSGVGDD134 pKa = 3.82PVSSDD139 pKa = 2.33ISGFDD144 pKa = 3.03WSVPQWALDD153 pKa = 3.27LDD155 pKa = 4.0ARR157 pKa = 11.84VRR159 pKa = 11.84AALSGAPTWLRR170 pKa = 11.84VHH172 pKa = 6.74EE173 pKa = 4.41KK174 pKa = 10.55RR175 pKa = 11.84AVCLGLSQLVFSDD188 pKa = 3.88GVVWDD193 pKa = 3.65QLEE196 pKa = 4.17PGIQKK201 pKa = 10.25SGSYY205 pKa = 8.29NTSSTNSRR213 pKa = 11.84IRR215 pKa = 11.84VMLAWLVARR224 pKa = 11.84RR225 pKa = 11.84IPQSRR230 pKa = 11.84YY231 pKa = 8.66GGRR234 pKa = 11.84DD235 pKa = 3.1ALEE238 pKa = 4.37GGGRR242 pKa = 11.84AMAMGDD248 pKa = 3.63DD249 pKa = 4.64AIEE252 pKa = 4.19TTTCMEE258 pKa = 4.25LLKK261 pKa = 10.69PNYY264 pKa = 9.06EE265 pKa = 3.84RR266 pKa = 11.84LGFVLKK272 pKa = 10.34EE273 pKa = 3.71VSRR276 pKa = 11.84DD277 pKa = 3.3IEE279 pKa = 4.06FCAYY283 pKa = 10.32AFDD286 pKa = 4.48LAQGYY291 pKa = 9.28RR292 pKa = 11.84PLRR295 pKa = 11.84VVKK298 pKa = 9.74MVAGLLRR305 pKa = 11.84TAVRR309 pKa = 11.84DD310 pKa = 3.71EE311 pKa = 3.85QHH313 pKa = 6.42EE314 pKa = 4.38EE315 pKa = 3.9EE316 pKa = 4.7LRR318 pKa = 11.84VALEE322 pKa = 3.79YY323 pKa = 10.63EE324 pKa = 4.38LRR326 pKa = 11.84HH327 pKa = 6.09SDD329 pKa = 3.0KK330 pKa = 10.96QAWAKK335 pKa = 10.26HH336 pKa = 5.15VIRR339 pKa = 11.84ASGWGARR346 pKa = 11.84KK347 pKa = 9.44
Molecular weight: 38.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEV9|A0A1L3KEV9_9VIRU Peptidase S39 domain-containing protein OS=Hubei sobemo-like virus 1 OX=1923194 PE=4 SV=1
MM1 pKa = 6.97VKK3 pKa = 10.35KK4 pKa = 10.42IRR6 pKa = 11.84VRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.0KK11 pKa = 9.93QAAKK15 pKa = 10.34APQPRR20 pKa = 11.84KK21 pKa = 10.07GVAQRR26 pKa = 11.84AMVQRR31 pKa = 11.84SVLDD35 pKa = 3.29SGAAAWARR43 pKa = 11.84LLADD47 pKa = 4.11PCAAPLTYY55 pKa = 9.82PCYY58 pKa = 8.87PTGTGGSVLMRR69 pKa = 11.84FEE71 pKa = 4.91SDD73 pKa = 3.51FLFATGATEE82 pKa = 3.71VAGIFGFCPGSLSGFSNATPLTSDD106 pKa = 2.67ILGTVLTLQSTSIPGFTFINANTNSYY132 pKa = 10.51RR133 pKa = 11.84PVAACMQVMYY143 pKa = 10.25PGTEE147 pKa = 4.05LNRR150 pKa = 11.84SGVVGVGICSGDD162 pKa = 3.37VLLRR166 pKa = 11.84NVNTVGGGSNINTNASEE183 pKa = 4.03VRR185 pKa = 11.84TMCQHH190 pKa = 5.71VEE192 pKa = 3.96RR193 pKa = 11.84MPTTVVEE200 pKa = 4.66IKK202 pKa = 9.57WFPGTADD209 pKa = 3.59EE210 pKa = 5.12EE211 pKa = 4.61NNSTQGLKK219 pKa = 10.68LGTLTDD225 pKa = 3.25IEE227 pKa = 4.32GRR229 pKa = 11.84NTIFMSASGFPVSTGIRR246 pKa = 11.84IRR248 pKa = 11.84MVAVYY253 pKa = 9.1EE254 pKa = 4.06LSFGAGAGQIAGSVAPTSGNTPAQVVRR281 pKa = 11.84ALTNRR286 pKa = 11.84DD287 pKa = 3.27ADD289 pKa = 3.83WYY291 pKa = 9.75ISSAAKK297 pKa = 9.82IGKK300 pKa = 9.43AVGNTISYY308 pKa = 10.04VATGYY313 pKa = 10.27KK314 pKa = 9.91AASAFAAGLALII326 pKa = 4.85
MM1 pKa = 6.97VKK3 pKa = 10.35KK4 pKa = 10.42IRR6 pKa = 11.84VRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.0KK11 pKa = 9.93QAAKK15 pKa = 10.34APQPRR20 pKa = 11.84KK21 pKa = 10.07GVAQRR26 pKa = 11.84AMVQRR31 pKa = 11.84SVLDD35 pKa = 3.29SGAAAWARR43 pKa = 11.84LLADD47 pKa = 4.11PCAAPLTYY55 pKa = 9.82PCYY58 pKa = 8.87PTGTGGSVLMRR69 pKa = 11.84FEE71 pKa = 4.91SDD73 pKa = 3.51FLFATGATEE82 pKa = 3.71VAGIFGFCPGSLSGFSNATPLTSDD106 pKa = 2.67ILGTVLTLQSTSIPGFTFINANTNSYY132 pKa = 10.51RR133 pKa = 11.84PVAACMQVMYY143 pKa = 10.25PGTEE147 pKa = 4.05LNRR150 pKa = 11.84SGVVGVGICSGDD162 pKa = 3.37VLLRR166 pKa = 11.84NVNTVGGGSNINTNASEE183 pKa = 4.03VRR185 pKa = 11.84TMCQHH190 pKa = 5.71VEE192 pKa = 3.96RR193 pKa = 11.84MPTTVVEE200 pKa = 4.66IKK202 pKa = 9.57WFPGTADD209 pKa = 3.59EE210 pKa = 5.12EE211 pKa = 4.61NNSTQGLKK219 pKa = 10.68LGTLTDD225 pKa = 3.25IEE227 pKa = 4.32GRR229 pKa = 11.84NTIFMSASGFPVSTGIRR246 pKa = 11.84IRR248 pKa = 11.84MVAVYY253 pKa = 9.1EE254 pKa = 4.06LSFGAGAGQIAGSVAPTSGNTPAQVVRR281 pKa = 11.84ALTNRR286 pKa = 11.84DD287 pKa = 3.27ADD289 pKa = 3.83WYY291 pKa = 9.75ISSAAKK297 pKa = 9.82IGKK300 pKa = 9.43AVGNTISYY308 pKa = 10.04VATGYY313 pKa = 10.27KK314 pKa = 9.91AASAFAAGLALII326 pKa = 4.85
Molecular weight: 34.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1210 |
326 |
537 |
403.3 |
43.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.0 ± 0.84 | 1.157 ± 0.254 |
4.132 ± 0.771 | 5.455 ± 1.214 |
3.554 ± 0.581 | 8.264 ± 0.918 |
1.322 ± 0.4 | 4.628 ± 0.349 |
4.628 ± 0.625 | 8.512 ± 1.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.479 ± 0.228 | 2.562 ± 0.899 |
5.702 ± 1.514 | 2.975 ± 0.442 |
6.116 ± 0.85 | 10.083 ± 1.265 |
5.372 ± 1.399 | 9.091 ± 0.21 |
1.24 ± 0.515 | 2.727 ± 0.232 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
