
Nitrosomonas aestuarii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Nitrosomonadaceae; Nitrosomonas
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
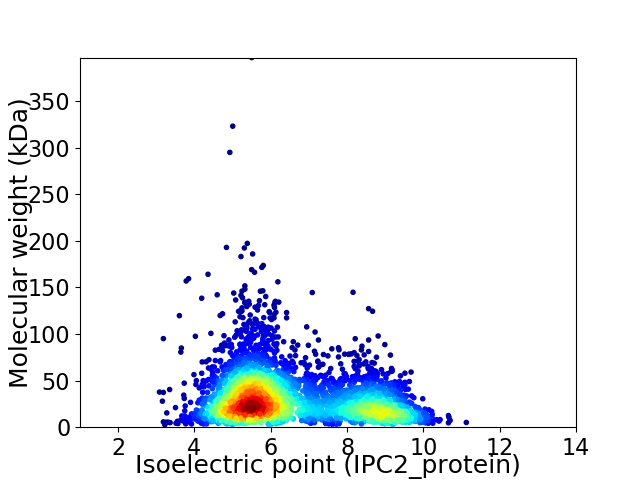
Virtual 2D-PAGE plot for 3744 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4DRV1|A0A1I4DRV1_9PROT Condensin subunit ScpA OS=Nitrosomonas aestuarii OX=52441 GN=SAMN05216302_102212 PE=3 SV=1
TT1 pKa = 7.26AGSTGGNTDD10 pKa = 3.0VNDD13 pKa = 4.63IISQITVNAGEE24 pKa = 4.33NSTDD28 pKa = 3.43NNFGEE33 pKa = 4.63VLPPPEE39 pKa = 4.6PASISGFVYY48 pKa = 10.71CDD50 pKa = 4.12DD51 pKa = 4.51NDD53 pKa = 5.49DD54 pKa = 4.6GIKK57 pKa = 10.65DD58 pKa = 3.38AGEE61 pKa = 4.41AGLSNVTIQLLNAAGVVINTTTTAADD87 pKa = 3.62GSYY90 pKa = 10.98SFTGLDD96 pKa = 2.95AGTYY100 pKa = 9.93SVVEE104 pKa = 4.18PTQPAGKK111 pKa = 9.91NDD113 pKa = 3.42GKK115 pKa = 9.34EE116 pKa = 4.19TAGSTGGNTDD126 pKa = 3.0VNDD129 pKa = 4.63IISQITVNAGEE140 pKa = 4.33NSTDD144 pKa = 3.41NNFGEE149 pKa = 4.68VTPTPVIDD157 pKa = 3.19IEE159 pKa = 4.36KK160 pKa = 9.82FVRR163 pKa = 11.84IEE165 pKa = 3.65QPAILFTGAVCEE177 pKa = 4.37VFGKK181 pKa = 9.33PLEE184 pKa = 4.09MTFDD188 pKa = 3.86YY189 pKa = 11.39NIGNTVDD196 pKa = 3.42TDD198 pKa = 3.54QDD200 pKa = 3.61SSKK203 pKa = 11.26AAILTQAGIEE213 pKa = 4.32DD214 pKa = 4.19DD215 pKa = 3.79GKK217 pKa = 11.34SYY219 pKa = 11.35VIVTDD224 pKa = 3.88KK225 pKa = 11.53SNPTDD230 pKa = 3.29SKK232 pKa = 11.54AKK234 pKa = 10.3VFFKK238 pKa = 10.92GYY240 pKa = 10.11VDD242 pKa = 3.6VGSEE246 pKa = 3.96FTASVNADD254 pKa = 2.9ADD256 pKa = 3.91KK257 pKa = 10.82FGSSTYY263 pKa = 9.59VHH265 pKa = 7.02FFDD268 pKa = 6.5DD269 pKa = 4.04NPFDD273 pKa = 3.96AVTGGDD279 pKa = 4.0ALLQSLTYY287 pKa = 8.88HH288 pKa = 6.46TSCSQPIHH296 pKa = 6.82LGDD299 pKa = 3.43MVGNVTLVDD308 pKa = 3.77YY309 pKa = 11.12VGEE312 pKa = 4.89DD313 pKa = 3.35GAAPDD318 pKa = 4.33SVPGVIGPDD327 pKa = 3.1EE328 pKa = 4.67DD329 pKa = 5.51ADD331 pKa = 4.2VPSGPAGEE339 pKa = 4.56TSVDD343 pKa = 3.53TAVFTYY349 pKa = 10.72YY350 pKa = 9.03VTNPGTVSLSNVVVTDD366 pKa = 3.58DD367 pKa = 4.51RR368 pKa = 11.84IAAIEE373 pKa = 4.6FIGGDD378 pKa = 3.87DD379 pKa = 4.27NNDD382 pKa = 3.33GLLDD386 pKa = 4.15PSEE389 pKa = 4.27TWSYY393 pKa = 8.88TASEE397 pKa = 4.19LVMNGLQTNIGTVTGAYY414 pKa = 10.76NNIQVTDD421 pKa = 4.07NDD423 pKa = 4.0PANYY427 pKa = 9.29IGMAAPPAINIEE439 pKa = 4.24KK440 pKa = 8.76YY441 pKa = 10.25VAVAGYY447 pKa = 10.4APAIEE452 pKa = 5.23HH453 pKa = 6.62ICDD456 pKa = 3.61TLGKK460 pKa = 8.12PLSMTFDD467 pKa = 3.7YY468 pKa = 11.23EE469 pKa = 3.84IGNTVDD475 pKa = 3.3TDD477 pKa = 3.54QDD479 pKa = 3.61SSKK482 pKa = 11.26AAILTQAGIEE492 pKa = 4.32DD493 pKa = 4.19DD494 pKa = 3.79GKK496 pKa = 11.19SYY498 pKa = 11.53VVVTDD503 pKa = 3.96KK504 pKa = 11.6SNPTDD509 pKa = 3.29SKK511 pKa = 11.54AKK513 pKa = 10.3VFFKK517 pKa = 10.92GYY519 pKa = 10.11VDD521 pKa = 3.6VGSEE525 pKa = 3.96FTASVNADD533 pKa = 2.93ADD535 pKa = 3.96KK536 pKa = 10.86FGSATYY542 pKa = 10.87VHH544 pKa = 7.04FFDD547 pKa = 6.41DD548 pKa = 4.04NPFDD552 pKa = 3.96AVTGGDD558 pKa = 4.0ALLQSLTYY566 pKa = 8.88HH567 pKa = 6.46TSCSQPINLGDD578 pKa = 3.62TVGNVSLVGYY588 pKa = 9.91AGEE591 pKa = 5.63DD592 pKa = 3.58GTIDD596 pKa = 3.41PSVWFGEE603 pKa = 4.51DD604 pKa = 3.15ADD606 pKa = 4.44TPSGPEE612 pKa = 3.6AAFGTAVDD620 pKa = 4.08FTYY623 pKa = 11.11VVTNPGSTPLANVSVSDD640 pKa = 4.1DD641 pKa = 3.32RR642 pKa = 11.84LSNVTYY648 pKa = 11.15VEE650 pKa = 4.32GDD652 pKa = 3.41TNADD656 pKa = 3.28NKK658 pKa = 10.66LDD660 pKa = 4.22PGEE663 pKa = 3.93AWIYY667 pKa = 7.79TASEE671 pKa = 4.06TALVGQVTNTGIVTATPVDD690 pKa = 3.5ALGNPLGLPEE700 pKa = 4.98VTDD703 pKa = 3.68QDD705 pKa = 3.72DD706 pKa = 3.39ANYY709 pKa = 9.9IVVGGPQIDD718 pKa = 3.51IEE720 pKa = 4.76KK721 pKa = 8.5YY722 pKa = 9.11VAVAGYY728 pKa = 10.29APAIGHH734 pKa = 6.4ICDD737 pKa = 3.74TLGKK741 pKa = 8.12PLSMTFDD748 pKa = 3.76YY749 pKa = 11.46NIGNTVDD756 pKa = 3.42TDD758 pKa = 3.54QDD760 pKa = 3.61SSKK763 pKa = 11.26AAILTQAGIEE773 pKa = 4.32DD774 pKa = 4.19DD775 pKa = 3.79GKK777 pKa = 11.34SYY779 pKa = 11.35VIVTDD784 pKa = 3.88KK785 pKa = 11.53SNPTDD790 pKa = 3.29SKK792 pKa = 11.54AKK794 pKa = 10.3VFFKK798 pKa = 10.92GYY800 pKa = 10.11VDD802 pKa = 3.6VGSEE806 pKa = 3.96FTASVNADD814 pKa = 2.9ADD816 pKa = 3.91KK817 pKa = 10.82FGSSTYY823 pKa = 9.59VHH825 pKa = 7.02FFDD828 pKa = 6.5DD829 pKa = 4.04NPFDD833 pKa = 3.96AVTGGDD839 pKa = 4.0ALLQSLTYY847 pKa = 8.88HH848 pKa = 6.46TSCSQPINLGDD859 pKa = 3.24IVGNVSLVGYY869 pKa = 9.91AGEE872 pKa = 5.63DD873 pKa = 3.58GTIDD877 pKa = 3.41PSVWFGEE884 pKa = 4.47DD885 pKa = 3.38ADD887 pKa = 4.53TGPGPQSLIGDD898 pKa = 3.99NVDD901 pKa = 3.69FTYY904 pKa = 11.13VVTNPGSTPLANVSLTDD921 pKa = 3.23NRR923 pKa = 11.84LNIAPGFEE931 pKa = 3.94GDD933 pKa = 3.51ANQNGLLDD941 pKa = 4.84PGEE944 pKa = 4.07EE945 pKa = 4.21WIYY948 pKa = 9.6TASEE952 pKa = 3.86LAEE955 pKa = 4.47AGQVTNIATVSAIAVDD971 pKa = 3.9AQGQDD976 pKa = 3.01LGLPSVSDD984 pKa = 3.39QDD986 pKa = 3.44AANYY990 pKa = 10.42NGVDD994 pKa = 3.66MLFPGYY1000 pKa = 9.41DD1001 pKa = 3.13ACDD1004 pKa = 3.31TLGKK1008 pKa = 9.77PKK1010 pKa = 10.38ALKK1013 pKa = 9.95FDD1015 pKa = 4.19YY1016 pKa = 10.64EE1017 pKa = 4.14PSVLVNTAQSSDD1029 pKa = 3.08KK1030 pKa = 11.33AKK1032 pKa = 10.52ILFDD1036 pKa = 4.04SGLVDD1041 pKa = 4.2DD1042 pKa = 5.84DD1043 pKa = 4.0GVSFILVTNKK1053 pKa = 10.2SNPTDD1058 pKa = 3.56LGGKK1062 pKa = 7.91EE1063 pKa = 4.01FFMGTVQVGDD1073 pKa = 3.79EE1074 pKa = 5.29FIASTTIAGDD1084 pKa = 3.35KK1085 pKa = 10.36FGSNTYY1091 pKa = 9.47VHH1093 pKa = 6.56VFDD1096 pKa = 6.58DD1097 pKa = 5.2NPFDD1101 pKa = 4.34TNGSNQLLQSAQYY1114 pKa = 8.24HH1115 pKa = 5.44TSCSQPIHH1123 pKa = 7.13LGDD1126 pKa = 3.59QLGDD1130 pKa = 3.57VTLVGYY1136 pKa = 10.21QGEE1139 pKa = 4.35SGAWGII1145 pKa = 4.66
TT1 pKa = 7.26AGSTGGNTDD10 pKa = 3.0VNDD13 pKa = 4.63IISQITVNAGEE24 pKa = 4.33NSTDD28 pKa = 3.43NNFGEE33 pKa = 4.63VLPPPEE39 pKa = 4.6PASISGFVYY48 pKa = 10.71CDD50 pKa = 4.12DD51 pKa = 4.51NDD53 pKa = 5.49DD54 pKa = 4.6GIKK57 pKa = 10.65DD58 pKa = 3.38AGEE61 pKa = 4.41AGLSNVTIQLLNAAGVVINTTTTAADD87 pKa = 3.62GSYY90 pKa = 10.98SFTGLDD96 pKa = 2.95AGTYY100 pKa = 9.93SVVEE104 pKa = 4.18PTQPAGKK111 pKa = 9.91NDD113 pKa = 3.42GKK115 pKa = 9.34EE116 pKa = 4.19TAGSTGGNTDD126 pKa = 3.0VNDD129 pKa = 4.63IISQITVNAGEE140 pKa = 4.33NSTDD144 pKa = 3.41NNFGEE149 pKa = 4.68VTPTPVIDD157 pKa = 3.19IEE159 pKa = 4.36KK160 pKa = 9.82FVRR163 pKa = 11.84IEE165 pKa = 3.65QPAILFTGAVCEE177 pKa = 4.37VFGKK181 pKa = 9.33PLEE184 pKa = 4.09MTFDD188 pKa = 3.86YY189 pKa = 11.39NIGNTVDD196 pKa = 3.42TDD198 pKa = 3.54QDD200 pKa = 3.61SSKK203 pKa = 11.26AAILTQAGIEE213 pKa = 4.32DD214 pKa = 4.19DD215 pKa = 3.79GKK217 pKa = 11.34SYY219 pKa = 11.35VIVTDD224 pKa = 3.88KK225 pKa = 11.53SNPTDD230 pKa = 3.29SKK232 pKa = 11.54AKK234 pKa = 10.3VFFKK238 pKa = 10.92GYY240 pKa = 10.11VDD242 pKa = 3.6VGSEE246 pKa = 3.96FTASVNADD254 pKa = 2.9ADD256 pKa = 3.91KK257 pKa = 10.82FGSSTYY263 pKa = 9.59VHH265 pKa = 7.02FFDD268 pKa = 6.5DD269 pKa = 4.04NPFDD273 pKa = 3.96AVTGGDD279 pKa = 4.0ALLQSLTYY287 pKa = 8.88HH288 pKa = 6.46TSCSQPIHH296 pKa = 6.82LGDD299 pKa = 3.43MVGNVTLVDD308 pKa = 3.77YY309 pKa = 11.12VGEE312 pKa = 4.89DD313 pKa = 3.35GAAPDD318 pKa = 4.33SVPGVIGPDD327 pKa = 3.1EE328 pKa = 4.67DD329 pKa = 5.51ADD331 pKa = 4.2VPSGPAGEE339 pKa = 4.56TSVDD343 pKa = 3.53TAVFTYY349 pKa = 10.72YY350 pKa = 9.03VTNPGTVSLSNVVVTDD366 pKa = 3.58DD367 pKa = 4.51RR368 pKa = 11.84IAAIEE373 pKa = 4.6FIGGDD378 pKa = 3.87DD379 pKa = 4.27NNDD382 pKa = 3.33GLLDD386 pKa = 4.15PSEE389 pKa = 4.27TWSYY393 pKa = 8.88TASEE397 pKa = 4.19LVMNGLQTNIGTVTGAYY414 pKa = 10.76NNIQVTDD421 pKa = 4.07NDD423 pKa = 4.0PANYY427 pKa = 9.29IGMAAPPAINIEE439 pKa = 4.24KK440 pKa = 8.76YY441 pKa = 10.25VAVAGYY447 pKa = 10.4APAIEE452 pKa = 5.23HH453 pKa = 6.62ICDD456 pKa = 3.61TLGKK460 pKa = 8.12PLSMTFDD467 pKa = 3.7YY468 pKa = 11.23EE469 pKa = 3.84IGNTVDD475 pKa = 3.3TDD477 pKa = 3.54QDD479 pKa = 3.61SSKK482 pKa = 11.26AAILTQAGIEE492 pKa = 4.32DD493 pKa = 4.19DD494 pKa = 3.79GKK496 pKa = 11.19SYY498 pKa = 11.53VVVTDD503 pKa = 3.96KK504 pKa = 11.6SNPTDD509 pKa = 3.29SKK511 pKa = 11.54AKK513 pKa = 10.3VFFKK517 pKa = 10.92GYY519 pKa = 10.11VDD521 pKa = 3.6VGSEE525 pKa = 3.96FTASVNADD533 pKa = 2.93ADD535 pKa = 3.96KK536 pKa = 10.86FGSATYY542 pKa = 10.87VHH544 pKa = 7.04FFDD547 pKa = 6.41DD548 pKa = 4.04NPFDD552 pKa = 3.96AVTGGDD558 pKa = 4.0ALLQSLTYY566 pKa = 8.88HH567 pKa = 6.46TSCSQPINLGDD578 pKa = 3.62TVGNVSLVGYY588 pKa = 9.91AGEE591 pKa = 5.63DD592 pKa = 3.58GTIDD596 pKa = 3.41PSVWFGEE603 pKa = 4.51DD604 pKa = 3.15ADD606 pKa = 4.44TPSGPEE612 pKa = 3.6AAFGTAVDD620 pKa = 4.08FTYY623 pKa = 11.11VVTNPGSTPLANVSVSDD640 pKa = 4.1DD641 pKa = 3.32RR642 pKa = 11.84LSNVTYY648 pKa = 11.15VEE650 pKa = 4.32GDD652 pKa = 3.41TNADD656 pKa = 3.28NKK658 pKa = 10.66LDD660 pKa = 4.22PGEE663 pKa = 3.93AWIYY667 pKa = 7.79TASEE671 pKa = 4.06TALVGQVTNTGIVTATPVDD690 pKa = 3.5ALGNPLGLPEE700 pKa = 4.98VTDD703 pKa = 3.68QDD705 pKa = 3.72DD706 pKa = 3.39ANYY709 pKa = 9.9IVVGGPQIDD718 pKa = 3.51IEE720 pKa = 4.76KK721 pKa = 8.5YY722 pKa = 9.11VAVAGYY728 pKa = 10.29APAIGHH734 pKa = 6.4ICDD737 pKa = 3.74TLGKK741 pKa = 8.12PLSMTFDD748 pKa = 3.76YY749 pKa = 11.46NIGNTVDD756 pKa = 3.42TDD758 pKa = 3.54QDD760 pKa = 3.61SSKK763 pKa = 11.26AAILTQAGIEE773 pKa = 4.32DD774 pKa = 4.19DD775 pKa = 3.79GKK777 pKa = 11.34SYY779 pKa = 11.35VIVTDD784 pKa = 3.88KK785 pKa = 11.53SNPTDD790 pKa = 3.29SKK792 pKa = 11.54AKK794 pKa = 10.3VFFKK798 pKa = 10.92GYY800 pKa = 10.11VDD802 pKa = 3.6VGSEE806 pKa = 3.96FTASVNADD814 pKa = 2.9ADD816 pKa = 3.91KK817 pKa = 10.82FGSSTYY823 pKa = 9.59VHH825 pKa = 7.02FFDD828 pKa = 6.5DD829 pKa = 4.04NPFDD833 pKa = 3.96AVTGGDD839 pKa = 4.0ALLQSLTYY847 pKa = 8.88HH848 pKa = 6.46TSCSQPINLGDD859 pKa = 3.24IVGNVSLVGYY869 pKa = 9.91AGEE872 pKa = 5.63DD873 pKa = 3.58GTIDD877 pKa = 3.41PSVWFGEE884 pKa = 4.47DD885 pKa = 3.38ADD887 pKa = 4.53TGPGPQSLIGDD898 pKa = 3.99NVDD901 pKa = 3.69FTYY904 pKa = 11.13VVTNPGSTPLANVSLTDD921 pKa = 3.23NRR923 pKa = 11.84LNIAPGFEE931 pKa = 3.94GDD933 pKa = 3.51ANQNGLLDD941 pKa = 4.84PGEE944 pKa = 4.07EE945 pKa = 4.21WIYY948 pKa = 9.6TASEE952 pKa = 3.86LAEE955 pKa = 4.47AGQVTNIATVSAIAVDD971 pKa = 3.9AQGQDD976 pKa = 3.01LGLPSVSDD984 pKa = 3.39QDD986 pKa = 3.44AANYY990 pKa = 10.42NGVDD994 pKa = 3.66MLFPGYY1000 pKa = 9.41DD1001 pKa = 3.13ACDD1004 pKa = 3.31TLGKK1008 pKa = 9.77PKK1010 pKa = 10.38ALKK1013 pKa = 9.95FDD1015 pKa = 4.19YY1016 pKa = 10.64EE1017 pKa = 4.14PSVLVNTAQSSDD1029 pKa = 3.08KK1030 pKa = 11.33AKK1032 pKa = 10.52ILFDD1036 pKa = 4.04SGLVDD1041 pKa = 4.2DD1042 pKa = 5.84DD1043 pKa = 4.0GVSFILVTNKK1053 pKa = 10.2SNPTDD1058 pKa = 3.56LGGKK1062 pKa = 7.91EE1063 pKa = 4.01FFMGTVQVGDD1073 pKa = 3.79EE1074 pKa = 5.29FIASTTIAGDD1084 pKa = 3.35KK1085 pKa = 10.36FGSNTYY1091 pKa = 9.47VHH1093 pKa = 6.56VFDD1096 pKa = 6.58DD1097 pKa = 5.2NPFDD1101 pKa = 4.34TNGSNQLLQSAQYY1114 pKa = 8.24HH1115 pKa = 5.44TSCSQPIHH1123 pKa = 7.13LGDD1126 pKa = 3.59QLGDD1130 pKa = 3.57VTLVGYY1136 pKa = 10.21QGEE1139 pKa = 4.35SGAWGII1145 pKa = 4.66
Molecular weight: 119.68 kDa
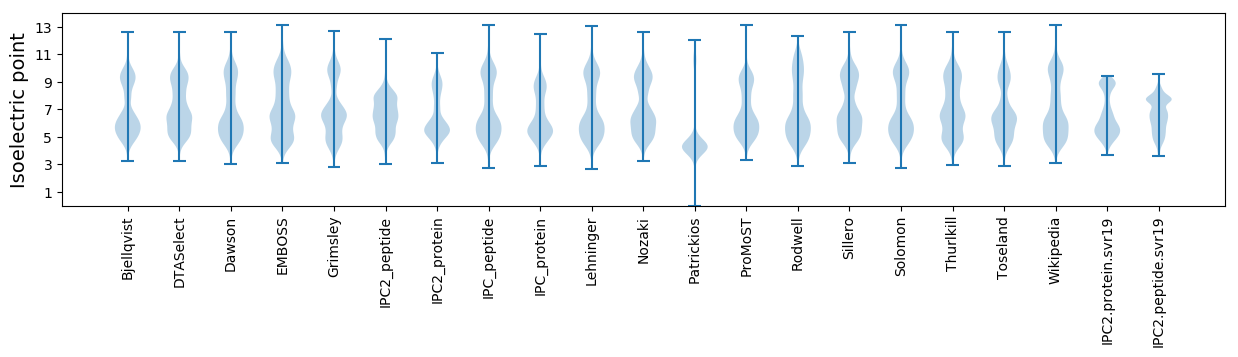
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3XQH5|A0A1I3XQH5_9PROT Uncharacterized protein OS=Nitrosomonas aestuarii OX=52441 GN=SAMN05216302_100215 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.14QPSVTKK11 pKa = 10.56RR12 pKa = 11.84KK13 pKa = 7.62RR14 pKa = 11.84THH16 pKa = 5.71GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 9.24TRR25 pKa = 11.84GGAAVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.96GRR39 pKa = 11.84ARR41 pKa = 11.84LSVV44 pKa = 3.12
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.14QPSVTKK11 pKa = 10.56RR12 pKa = 11.84KK13 pKa = 7.62RR14 pKa = 11.84THH16 pKa = 5.71GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 9.24TRR25 pKa = 11.84GGAAVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.96GRR39 pKa = 11.84ARR41 pKa = 11.84LSVV44 pKa = 3.12
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1122864 |
25 |
3466 |
299.9 |
33.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.607 ± 0.041 | 1.03 ± 0.013 |
5.444 ± 0.031 | 5.972 ± 0.039 |
4.15 ± 0.03 | 6.688 ± 0.044 |
2.508 ± 0.023 | 7.014 ± 0.032 |
5.071 ± 0.036 | 10.216 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.526 ± 0.02 | 4.459 ± 0.036 |
4.165 ± 0.023 | 4.344 ± 0.033 |
5.341 ± 0.035 | 6.226 ± 0.034 |
5.519 ± 0.033 | 6.463 ± 0.031 |
1.273 ± 0.02 | 2.984 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
