
bacterium HR12
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
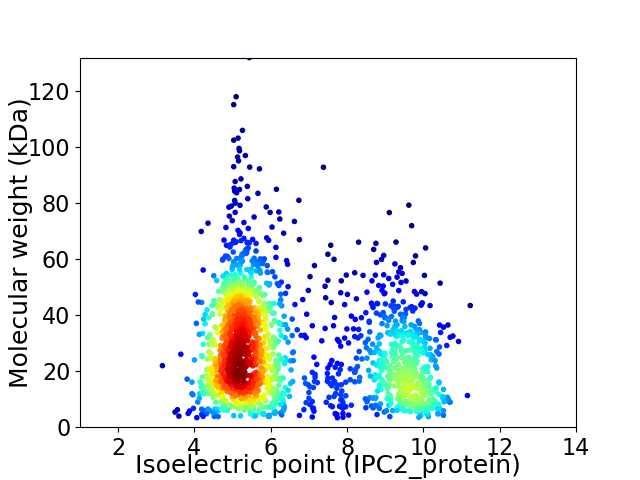
Virtual 2D-PAGE plot for 1868 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5WE58|A0A2H5WE58_9BACT Purine catabolism regulatory protein OS=bacterium HR12 OX=2035407 GN=pucR_2 PE=3 SV=1
MM1 pKa = 7.59PVGAEE6 pKa = 4.1NPPGALMLMDD16 pKa = 4.64WYY18 pKa = 10.07YY19 pKa = 10.44QPKK22 pKa = 7.96IAAMVTEE29 pKa = 4.55WVLYY33 pKa = 9.59LSPCKK38 pKa = 10.15GVRR41 pKa = 11.84EE42 pKa = 4.3VILTDD47 pKa = 3.65AEE49 pKa = 4.09QALEE53 pKa = 4.37DD54 pKa = 4.57GYY56 pKa = 11.49KK57 pKa = 10.62GYY59 pKa = 11.32ANKK62 pKa = 10.5LYY64 pKa = 8.85QTAEE68 pKa = 4.08AEE70 pKa = 4.4VAFPSDD76 pKa = 3.54EE77 pKa = 4.22TLSLAEE83 pKa = 4.43FGTNITTDD91 pKa = 3.31EE92 pKa = 4.19QAQEE96 pKa = 3.37WDD98 pKa = 4.49AIFLPISQQQ107 pKa = 2.99
MM1 pKa = 7.59PVGAEE6 pKa = 4.1NPPGALMLMDD16 pKa = 4.64WYY18 pKa = 10.07YY19 pKa = 10.44QPKK22 pKa = 7.96IAAMVTEE29 pKa = 4.55WVLYY33 pKa = 9.59LSPCKK38 pKa = 10.15GVRR41 pKa = 11.84EE42 pKa = 4.3VILTDD47 pKa = 3.65AEE49 pKa = 4.09QALEE53 pKa = 4.37DD54 pKa = 4.57GYY56 pKa = 11.49KK57 pKa = 10.62GYY59 pKa = 11.32ANKK62 pKa = 10.5LYY64 pKa = 8.85QTAEE68 pKa = 4.08AEE70 pKa = 4.4VAFPSDD76 pKa = 3.54EE77 pKa = 4.22TLSLAEE83 pKa = 4.43FGTNITTDD91 pKa = 3.31EE92 pKa = 4.19QAQEE96 pKa = 3.37WDD98 pKa = 4.49AIFLPISQQQ107 pKa = 2.99
Molecular weight: 11.94 kDa
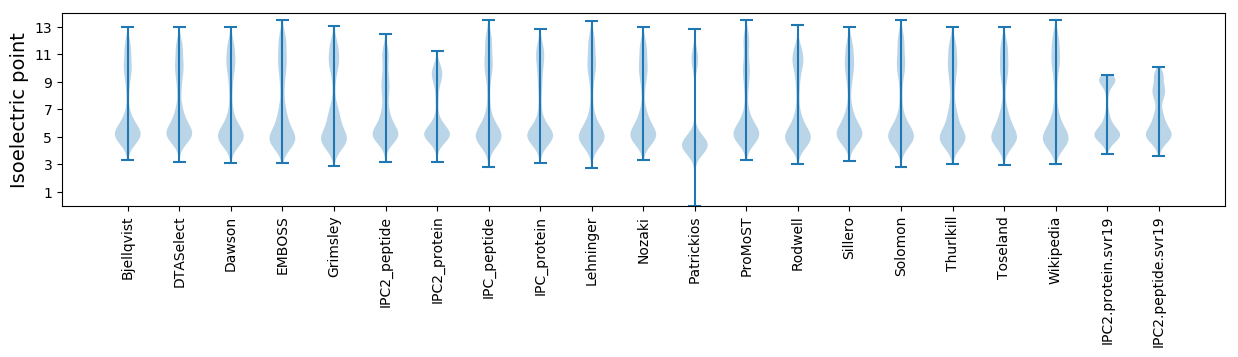
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5WEW4|A0A2H5WEW4_9BACT Imidazolonepropionase OS=bacterium HR12 OX=2035407 GN=hutI_3 PE=4 SV=1
MM1 pKa = 7.3AVEE4 pKa = 4.17EE5 pKa = 4.05ARR7 pKa = 11.84RR8 pKa = 11.84AGVGRR13 pKa = 11.84LGGRR17 pKa = 11.84GGGLEE22 pKa = 3.95QGRR25 pKa = 11.84DD26 pKa = 3.47EE27 pKa = 4.98GPPSGRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AEE38 pKa = 3.85APQGPRR44 pKa = 11.84AGSRR48 pKa = 11.84AGAPGRR54 pKa = 11.84AARR57 pKa = 11.84AGPGAARR64 pKa = 11.84ACRR67 pKa = 11.84SSPRR71 pKa = 11.84PPTPGPTGPAAPSAPRR87 pKa = 11.84GRR89 pKa = 11.84RR90 pKa = 11.84PPAGARR96 pKa = 11.84ARR98 pKa = 11.84RR99 pKa = 11.84TRR101 pKa = 11.84RR102 pKa = 11.84GSRR105 pKa = 11.84GARR108 pKa = 11.84GAPGGSRR115 pKa = 11.84PRR117 pKa = 11.84VARR120 pKa = 11.84SAPGSGSSPRR130 pKa = 11.84RR131 pKa = 11.84GTRR134 pKa = 11.84ARR136 pKa = 11.84SVRR139 pKa = 11.84PPVRR143 pKa = 11.84SHH145 pKa = 7.09PSRR148 pKa = 11.84PTRR151 pKa = 11.84PPRR154 pKa = 11.84PQGARR159 pKa = 11.84PGPPGGSPAPGRR171 pKa = 11.84GARR174 pKa = 11.84RR175 pKa = 11.84SRR177 pKa = 11.84GSEE180 pKa = 3.92RR181 pKa = 11.84GWGRR185 pKa = 11.84PGGARR190 pKa = 11.84SPRR193 pKa = 11.84RR194 pKa = 11.84ARR196 pKa = 11.84PAPASGPRR204 pKa = 11.84GPARR208 pKa = 11.84RR209 pKa = 11.84GGTGRR214 pKa = 11.84GPPRR218 pKa = 11.84PPAPPRR224 pKa = 11.84AGGRR228 pKa = 11.84RR229 pKa = 11.84GSGAAAVGGRR239 pKa = 11.84RR240 pKa = 11.84SRR242 pKa = 11.84TTPSPGLPAGSRR254 pKa = 11.84RR255 pKa = 11.84GRR257 pKa = 11.84PARR260 pKa = 11.84RR261 pKa = 11.84AQARR265 pKa = 11.84PGPRR269 pKa = 11.84GRR271 pKa = 11.84VRR273 pKa = 11.84GAGRR277 pKa = 11.84GARR280 pKa = 11.84PRR282 pKa = 11.84RR283 pKa = 11.84WRR285 pKa = 11.84GTGRR289 pKa = 11.84GSARR293 pKa = 11.84SGGRR297 pKa = 11.84ARR299 pKa = 11.84ARR301 pKa = 11.84RR302 pKa = 11.84PARR305 pKa = 11.84AWCTRR310 pKa = 11.84SAGRR314 pKa = 11.84AGSARR319 pKa = 11.84RR320 pKa = 11.84ARR322 pKa = 11.84PAAARR327 pKa = 11.84RR328 pKa = 11.84PPRR331 pKa = 11.84RR332 pKa = 11.84RR333 pKa = 11.84GAPRR337 pKa = 11.84PRR339 pKa = 11.84RR340 pKa = 11.84GPGSRR345 pKa = 11.84VPRR348 pKa = 11.84APARR352 pKa = 11.84RR353 pKa = 11.84RR354 pKa = 11.84GLRR357 pKa = 11.84RR358 pKa = 11.84RR359 pKa = 11.84RR360 pKa = 11.84RR361 pKa = 11.84SGPRR365 pKa = 11.84APRR368 pKa = 11.84ARR370 pKa = 11.84GPRR373 pKa = 11.84PAGAGRR379 pKa = 11.84GGRR382 pKa = 11.84RR383 pKa = 11.84RR384 pKa = 11.84SAPAVPRR391 pKa = 11.84RR392 pKa = 11.84APGARR397 pKa = 11.84ARR399 pKa = 11.84ARR401 pKa = 11.84RR402 pKa = 11.84WPRR405 pKa = 11.84DD406 pKa = 3.2PGAPAGPRR414 pKa = 11.84SRR416 pKa = 11.84GARR419 pKa = 3.36
MM1 pKa = 7.3AVEE4 pKa = 4.17EE5 pKa = 4.05ARR7 pKa = 11.84RR8 pKa = 11.84AGVGRR13 pKa = 11.84LGGRR17 pKa = 11.84GGGLEE22 pKa = 3.95QGRR25 pKa = 11.84DD26 pKa = 3.47EE27 pKa = 4.98GPPSGRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AEE38 pKa = 3.85APQGPRR44 pKa = 11.84AGSRR48 pKa = 11.84AGAPGRR54 pKa = 11.84AARR57 pKa = 11.84AGPGAARR64 pKa = 11.84ACRR67 pKa = 11.84SSPRR71 pKa = 11.84PPTPGPTGPAAPSAPRR87 pKa = 11.84GRR89 pKa = 11.84RR90 pKa = 11.84PPAGARR96 pKa = 11.84ARR98 pKa = 11.84RR99 pKa = 11.84TRR101 pKa = 11.84RR102 pKa = 11.84GSRR105 pKa = 11.84GARR108 pKa = 11.84GAPGGSRR115 pKa = 11.84PRR117 pKa = 11.84VARR120 pKa = 11.84SAPGSGSSPRR130 pKa = 11.84RR131 pKa = 11.84GTRR134 pKa = 11.84ARR136 pKa = 11.84SVRR139 pKa = 11.84PPVRR143 pKa = 11.84SHH145 pKa = 7.09PSRR148 pKa = 11.84PTRR151 pKa = 11.84PPRR154 pKa = 11.84PQGARR159 pKa = 11.84PGPPGGSPAPGRR171 pKa = 11.84GARR174 pKa = 11.84RR175 pKa = 11.84SRR177 pKa = 11.84GSEE180 pKa = 3.92RR181 pKa = 11.84GWGRR185 pKa = 11.84PGGARR190 pKa = 11.84SPRR193 pKa = 11.84RR194 pKa = 11.84ARR196 pKa = 11.84PAPASGPRR204 pKa = 11.84GPARR208 pKa = 11.84RR209 pKa = 11.84GGTGRR214 pKa = 11.84GPPRR218 pKa = 11.84PPAPPRR224 pKa = 11.84AGGRR228 pKa = 11.84RR229 pKa = 11.84GSGAAAVGGRR239 pKa = 11.84RR240 pKa = 11.84SRR242 pKa = 11.84TTPSPGLPAGSRR254 pKa = 11.84RR255 pKa = 11.84GRR257 pKa = 11.84PARR260 pKa = 11.84RR261 pKa = 11.84AQARR265 pKa = 11.84PGPRR269 pKa = 11.84GRR271 pKa = 11.84VRR273 pKa = 11.84GAGRR277 pKa = 11.84GARR280 pKa = 11.84PRR282 pKa = 11.84RR283 pKa = 11.84WRR285 pKa = 11.84GTGRR289 pKa = 11.84GSARR293 pKa = 11.84SGGRR297 pKa = 11.84ARR299 pKa = 11.84ARR301 pKa = 11.84RR302 pKa = 11.84PARR305 pKa = 11.84AWCTRR310 pKa = 11.84SAGRR314 pKa = 11.84AGSARR319 pKa = 11.84RR320 pKa = 11.84ARR322 pKa = 11.84PAAARR327 pKa = 11.84RR328 pKa = 11.84PPRR331 pKa = 11.84RR332 pKa = 11.84RR333 pKa = 11.84GAPRR337 pKa = 11.84PRR339 pKa = 11.84RR340 pKa = 11.84GPGSRR345 pKa = 11.84VPRR348 pKa = 11.84APARR352 pKa = 11.84RR353 pKa = 11.84RR354 pKa = 11.84GLRR357 pKa = 11.84RR358 pKa = 11.84RR359 pKa = 11.84RR360 pKa = 11.84RR361 pKa = 11.84SGPRR365 pKa = 11.84APRR368 pKa = 11.84ARR370 pKa = 11.84GPRR373 pKa = 11.84PAGAGRR379 pKa = 11.84GGRR382 pKa = 11.84RR383 pKa = 11.84RR384 pKa = 11.84SAPAVPRR391 pKa = 11.84RR392 pKa = 11.84APGARR397 pKa = 11.84ARR399 pKa = 11.84ARR401 pKa = 11.84RR402 pKa = 11.84WPRR405 pKa = 11.84DD406 pKa = 3.2PGAPAGPRR414 pKa = 11.84SRR416 pKa = 11.84GARR419 pKa = 3.36
Molecular weight: 43.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
496997 |
29 |
1205 |
266.1 |
28.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.115 ± 0.091 | 0.831 ± 0.019 |
5.148 ± 0.039 | 7.58 ± 0.066 |
2.942 ± 0.035 | 9.456 ± 0.067 |
1.97 ± 0.03 | 3.794 ± 0.051 |
1.572 ± 0.039 | 10.515 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.83 ± 0.026 | 1.391 ± 0.029 |
6.412 ± 0.059 | 2.039 ± 0.028 |
9.822 ± 0.085 | 4.237 ± 0.042 |
4.801 ± 0.045 | 9.136 ± 0.061 |
1.443 ± 0.028 | 1.968 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
