
Sewage-associated gemycircularvirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykrogvirus; Gemykrogvirus sewopo1; Sewage derived gemykrogvirus 1
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
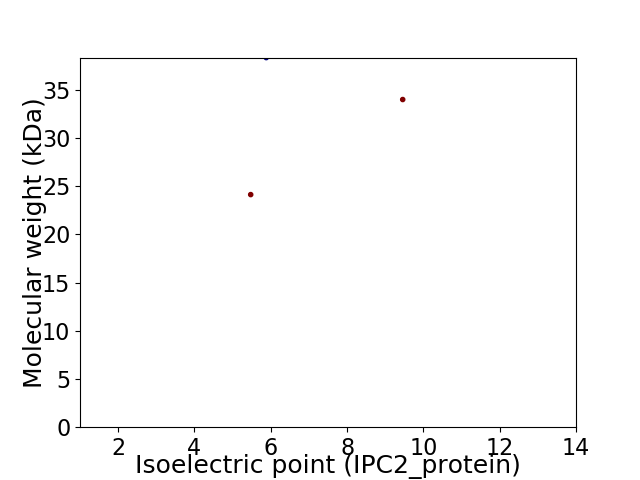
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
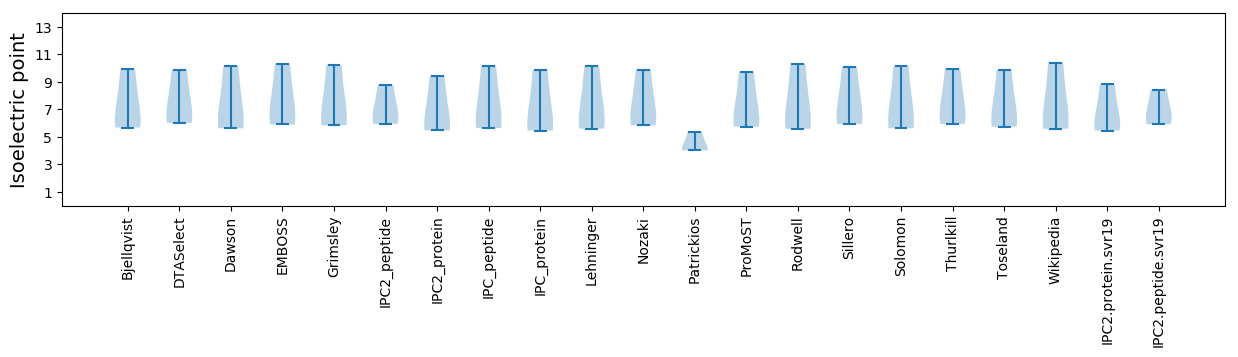
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7CL85|A0A0A7CL85_9VIRU RepA OS=Sewage-associated gemycircularvirus 4 OX=1519399 PE=3 SV=1
MM1 pKa = 7.68PRR3 pKa = 11.84FQARR7 pKa = 11.84AKK9 pKa = 10.39AFIVTFPQVSEE20 pKa = 4.81DD21 pKa = 3.29VQQRR25 pKa = 11.84FDD27 pKa = 3.79HH28 pKa = 6.59EE29 pKa = 4.66GASLLLNITQDD40 pKa = 4.22LKK42 pKa = 11.42DD43 pKa = 3.81PSCFRR48 pKa = 11.84LGRR51 pKa = 11.84EE52 pKa = 3.66RR53 pKa = 11.84HH54 pKa = 4.95QDD56 pKa = 3.28GGVHH60 pKa = 4.27YY61 pKa = 10.47HH62 pKa = 6.16MYY64 pKa = 10.16IGFDD68 pKa = 3.29EE69 pKa = 4.52VVHH72 pKa = 6.45INRR75 pKa = 11.84ANLFDD80 pKa = 3.8YY81 pKa = 10.9FGAHH85 pKa = 6.22GNIKK89 pKa = 9.47SVRR92 pKa = 11.84RR93 pKa = 11.84TPRR96 pKa = 11.84TVYY99 pKa = 10.19DD100 pKa = 3.55YY101 pKa = 10.92CGKK104 pKa = 10.53DD105 pKa = 2.73GDD107 pKa = 3.92VRR109 pKa = 11.84YY110 pKa = 10.08EE111 pKa = 3.96RR112 pKa = 11.84GDD114 pKa = 3.43PPEE117 pKa = 4.17SVSRR121 pKa = 11.84SRR123 pKa = 11.84GEE125 pKa = 4.33NGEE128 pKa = 4.1KK129 pKa = 9.32WHH131 pKa = 6.76TICDD135 pKa = 3.92APDD138 pKa = 3.8KK139 pKa = 10.79DD140 pKa = 4.34TFLSLCRR147 pKa = 11.84QLAPKK152 pKa = 10.2DD153 pKa = 3.39WLLSNSRR160 pKa = 11.84ILEE163 pKa = 4.18YY164 pKa = 11.17ANTYY168 pKa = 9.26YY169 pKa = 10.32PEE171 pKa = 4.74IPSPYY176 pKa = 9.02EE177 pKa = 3.76GPPIIAEE184 pKa = 3.92MEE186 pKa = 4.17RR187 pKa = 11.84YY188 pKa = 9.5PEE190 pKa = 4.05LEE192 pKa = 3.7RR193 pKa = 11.84WLEE196 pKa = 3.73QARR199 pKa = 11.84IGDD202 pKa = 4.02DD203 pKa = 3.28RR204 pKa = 11.84PEE206 pKa = 3.72RR207 pKa = 4.64
MM1 pKa = 7.68PRR3 pKa = 11.84FQARR7 pKa = 11.84AKK9 pKa = 10.39AFIVTFPQVSEE20 pKa = 4.81DD21 pKa = 3.29VQQRR25 pKa = 11.84FDD27 pKa = 3.79HH28 pKa = 6.59EE29 pKa = 4.66GASLLLNITQDD40 pKa = 4.22LKK42 pKa = 11.42DD43 pKa = 3.81PSCFRR48 pKa = 11.84LGRR51 pKa = 11.84EE52 pKa = 3.66RR53 pKa = 11.84HH54 pKa = 4.95QDD56 pKa = 3.28GGVHH60 pKa = 4.27YY61 pKa = 10.47HH62 pKa = 6.16MYY64 pKa = 10.16IGFDD68 pKa = 3.29EE69 pKa = 4.52VVHH72 pKa = 6.45INRR75 pKa = 11.84ANLFDD80 pKa = 3.8YY81 pKa = 10.9FGAHH85 pKa = 6.22GNIKK89 pKa = 9.47SVRR92 pKa = 11.84RR93 pKa = 11.84TPRR96 pKa = 11.84TVYY99 pKa = 10.19DD100 pKa = 3.55YY101 pKa = 10.92CGKK104 pKa = 10.53DD105 pKa = 2.73GDD107 pKa = 3.92VRR109 pKa = 11.84YY110 pKa = 10.08EE111 pKa = 3.96RR112 pKa = 11.84GDD114 pKa = 3.43PPEE117 pKa = 4.17SVSRR121 pKa = 11.84SRR123 pKa = 11.84GEE125 pKa = 4.33NGEE128 pKa = 4.1KK129 pKa = 9.32WHH131 pKa = 6.76TICDD135 pKa = 3.92APDD138 pKa = 3.8KK139 pKa = 10.79DD140 pKa = 4.34TFLSLCRR147 pKa = 11.84QLAPKK152 pKa = 10.2DD153 pKa = 3.39WLLSNSRR160 pKa = 11.84ILEE163 pKa = 4.18YY164 pKa = 11.17ANTYY168 pKa = 9.26YY169 pKa = 10.32PEE171 pKa = 4.74IPSPYY176 pKa = 9.02EE177 pKa = 3.76GPPIIAEE184 pKa = 3.92MEE186 pKa = 4.17RR187 pKa = 11.84YY188 pKa = 9.5PEE190 pKa = 4.05LEE192 pKa = 3.7RR193 pKa = 11.84WLEE196 pKa = 3.73QARR199 pKa = 11.84IGDD202 pKa = 4.02DD203 pKa = 3.28RR204 pKa = 11.84PEE206 pKa = 3.72RR207 pKa = 4.64
Molecular weight: 24.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7CL63|A0A0A7CL63_9VIRU Replication-associated protein OS=Sewage-associated gemycircularvirus 4 OX=1519399 PE=3 SV=1
MM1 pKa = 7.37ARR3 pKa = 11.84SFKK6 pKa = 9.99PRR8 pKa = 11.84YY9 pKa = 8.83RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84FSTRR16 pKa = 11.84RR17 pKa = 11.84PSFRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 7.01TYY25 pKa = 9.59RR26 pKa = 11.84GRR28 pKa = 11.84RR29 pKa = 11.84YY30 pKa = 9.6ARR32 pKa = 11.84RR33 pKa = 11.84PKK35 pKa = 10.15RR36 pKa = 11.84SRR38 pKa = 11.84VTPRR42 pKa = 11.84KK43 pKa = 9.3VRR45 pKa = 11.84NIASKK50 pKa = 10.55KK51 pKa = 9.86KK52 pKa = 9.85QDD54 pKa = 3.54TLLGSDD60 pKa = 4.39SSDD63 pKa = 3.26PDD65 pKa = 3.63DD66 pKa = 4.45FSNVVALTTGNNYY79 pKa = 9.53FLWTPTFRR87 pKa = 11.84DD88 pKa = 3.15RR89 pKa = 11.84DD90 pKa = 4.11DD91 pKa = 3.69NVNNHH96 pKa = 4.88TRR98 pKa = 11.84GAASCYY104 pKa = 9.12YY105 pKa = 9.99RR106 pKa = 11.84GVRR109 pKa = 11.84DD110 pKa = 4.25RR111 pKa = 11.84IMVAASFQLIHH122 pKa = 6.85RR123 pKa = 11.84RR124 pKa = 11.84VCFWSHH130 pKa = 4.88SQISSATPYY139 pKa = 10.25DD140 pKa = 3.45IPGDD144 pKa = 4.25DD145 pKa = 4.24GDD147 pKa = 3.31IAYY150 pKa = 7.77TRR152 pKa = 11.84RR153 pKa = 11.84PLQKK157 pKa = 9.91INPSTDD163 pKa = 2.97TATFRR168 pKa = 11.84YY169 pKa = 9.32LFKK172 pKa = 10.6GTVGVDD178 pKa = 3.75FTEE181 pKa = 4.58DD182 pKa = 3.75SRR184 pKa = 11.84WDD186 pKa = 3.36SPMANNRR193 pKa = 11.84IQVVYY198 pKa = 10.6DD199 pKa = 3.1RR200 pKa = 11.84QYY202 pKa = 11.26PVNPNYY208 pKa = 10.55NVAEE212 pKa = 4.33GNAFGKK218 pKa = 10.44ISTKK222 pKa = 10.66KK223 pKa = 9.61LWHH226 pKa = 6.61PMNSTVKK233 pKa = 10.58YY234 pKa = 10.47DD235 pKa = 3.15DD236 pKa = 3.64WEE238 pKa = 3.98QGRR241 pKa = 11.84EE242 pKa = 3.44IDD244 pKa = 3.72AASWGNAHH252 pKa = 6.96PASRR256 pKa = 11.84GNFYY260 pKa = 10.57ILDD263 pKa = 3.9MFSTGQDD270 pKa = 3.45VPSEE274 pKa = 4.29SATQCGSYY282 pKa = 8.87STEE285 pKa = 3.78STVYY289 pKa = 8.67WHH291 pKa = 6.9EE292 pKa = 4.22SSS294 pKa = 3.93
MM1 pKa = 7.37ARR3 pKa = 11.84SFKK6 pKa = 9.99PRR8 pKa = 11.84YY9 pKa = 8.83RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84FSTRR16 pKa = 11.84RR17 pKa = 11.84PSFRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 7.01TYY25 pKa = 9.59RR26 pKa = 11.84GRR28 pKa = 11.84RR29 pKa = 11.84YY30 pKa = 9.6ARR32 pKa = 11.84RR33 pKa = 11.84PKK35 pKa = 10.15RR36 pKa = 11.84SRR38 pKa = 11.84VTPRR42 pKa = 11.84KK43 pKa = 9.3VRR45 pKa = 11.84NIASKK50 pKa = 10.55KK51 pKa = 9.86KK52 pKa = 9.85QDD54 pKa = 3.54TLLGSDD60 pKa = 4.39SSDD63 pKa = 3.26PDD65 pKa = 3.63DD66 pKa = 4.45FSNVVALTTGNNYY79 pKa = 9.53FLWTPTFRR87 pKa = 11.84DD88 pKa = 3.15RR89 pKa = 11.84DD90 pKa = 4.11DD91 pKa = 3.69NVNNHH96 pKa = 4.88TRR98 pKa = 11.84GAASCYY104 pKa = 9.12YY105 pKa = 9.99RR106 pKa = 11.84GVRR109 pKa = 11.84DD110 pKa = 4.25RR111 pKa = 11.84IMVAASFQLIHH122 pKa = 6.85RR123 pKa = 11.84RR124 pKa = 11.84VCFWSHH130 pKa = 4.88SQISSATPYY139 pKa = 10.25DD140 pKa = 3.45IPGDD144 pKa = 4.25DD145 pKa = 4.24GDD147 pKa = 3.31IAYY150 pKa = 7.77TRR152 pKa = 11.84RR153 pKa = 11.84PLQKK157 pKa = 9.91INPSTDD163 pKa = 2.97TATFRR168 pKa = 11.84YY169 pKa = 9.32LFKK172 pKa = 10.6GTVGVDD178 pKa = 3.75FTEE181 pKa = 4.58DD182 pKa = 3.75SRR184 pKa = 11.84WDD186 pKa = 3.36SPMANNRR193 pKa = 11.84IQVVYY198 pKa = 10.6DD199 pKa = 3.1RR200 pKa = 11.84QYY202 pKa = 11.26PVNPNYY208 pKa = 10.55NVAEE212 pKa = 4.33GNAFGKK218 pKa = 10.44ISTKK222 pKa = 10.66KK223 pKa = 9.61LWHH226 pKa = 6.61PMNSTVKK233 pKa = 10.58YY234 pKa = 10.47DD235 pKa = 3.15DD236 pKa = 3.64WEE238 pKa = 3.98QGRR241 pKa = 11.84EE242 pKa = 3.44IDD244 pKa = 3.72AASWGNAHH252 pKa = 6.96PASRR256 pKa = 11.84GNFYY260 pKa = 10.57ILDD263 pKa = 3.9MFSTGQDD270 pKa = 3.45VPSEE274 pKa = 4.29SATQCGSYY282 pKa = 8.87STEE285 pKa = 3.78STVYY289 pKa = 8.67WHH291 pKa = 6.9EE292 pKa = 4.22SSS294 pKa = 3.93
Molecular weight: 34.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
831 |
207 |
330 |
277.0 |
32.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.415 ± 0.534 | 1.685 ± 0.328 |
7.702 ± 0.217 | 5.656 ± 1.658 |
4.934 ± 0.259 | 6.619 ± 0.597 |
2.768 ± 0.372 | 4.934 ± 0.59 |
4.573 ± 0.47 | 5.897 ± 1.404 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.564 ± 0.07 | 4.452 ± 0.676 |
6.017 ± 0.455 | 3.49 ± 0.221 |
9.627 ± 1.084 | 7.1 ± 1.534 |
4.934 ± 1.263 | 5.295 ± 0.256 |
2.046 ± 0.232 | 5.295 ± 0.079 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
