
Mouse mammary tumor virus (strain C3H) (MMTV)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Betaretrovirus; Mouse mammary tumor virus
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
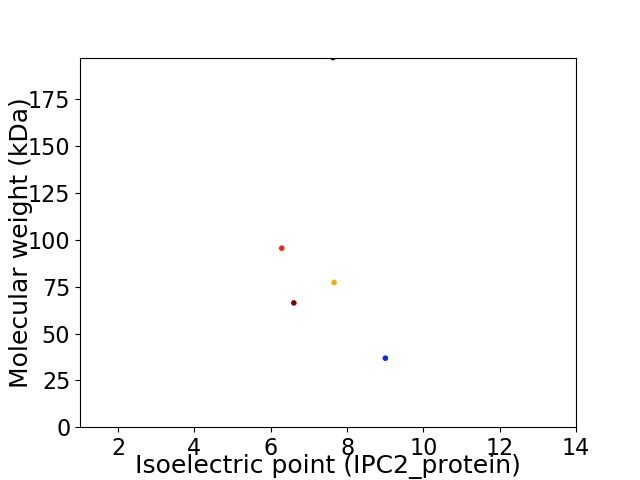
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9IZT2|PRO_MMTVC Gag-Pro polyprotein OS=Mouse mammary tumor virus (strain C3H) OX=11759 GN=gag-pro PE=1 SV=1
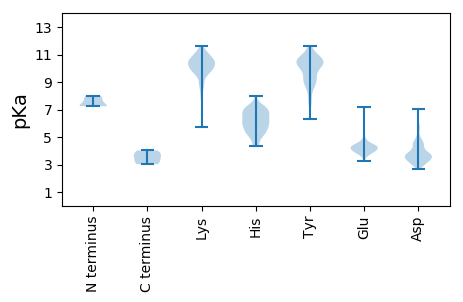
MM1 pKa = 7.24GVSGSKK7 pKa = 9.48GQKK10 pKa = 10.01LFVSVLQRR18 pKa = 11.84LLSEE22 pKa = 4.7RR23 pKa = 11.84GLHH26 pKa = 5.31VKK28 pKa = 10.05EE29 pKa = 4.39SSAIEE34 pKa = 4.43FYY36 pKa = 11.02QFLIKK41 pKa = 10.55VSPWFPEE48 pKa = 3.79EE49 pKa = 4.5GGLNLQDD56 pKa = 3.26WKK58 pKa = 11.05RR59 pKa = 11.84VGRR62 pKa = 11.84EE63 pKa = 3.7MKK65 pKa = 10.24KK66 pKa = 9.98YY67 pKa = 9.71AAEE70 pKa = 4.67HH71 pKa = 5.81GTDD74 pKa = 4.27SIPKK78 pKa = 7.84QAYY81 pKa = 7.84PIWLQLRR88 pKa = 11.84EE89 pKa = 4.17ILTEE93 pKa = 3.88QSDD96 pKa = 4.44LVLLSAEE103 pKa = 4.15AKK105 pKa = 10.18SVTEE109 pKa = 4.08EE110 pKa = 3.97EE111 pKa = 4.47LEE113 pKa = 4.17EE114 pKa = 4.46GLTGLLSASSQEE126 pKa = 3.85KK127 pKa = 9.34TYY129 pKa = 8.97GTRR132 pKa = 11.84GTAYY136 pKa = 10.45AEE138 pKa = 4.13IDD140 pKa = 3.78TEE142 pKa = 4.13VDD144 pKa = 3.01KK145 pKa = 11.4LSEE148 pKa = 4.53HH149 pKa = 7.04IYY151 pKa = 10.55DD152 pKa = 3.66EE153 pKa = 4.71PYY155 pKa = 10.62EE156 pKa = 4.21EE157 pKa = 5.61KK158 pKa = 10.79EE159 pKa = 4.01KK160 pKa = 10.96ADD162 pKa = 3.5KK163 pKa = 11.34NEE165 pKa = 4.22EE166 pKa = 3.82KK167 pKa = 11.02DD168 pKa = 3.3HH169 pKa = 5.17VRR171 pKa = 11.84KK172 pKa = 9.7VKK174 pKa = 10.63KK175 pKa = 9.75IVQRR179 pKa = 11.84KK180 pKa = 7.92EE181 pKa = 3.59NSEE184 pKa = 4.13HH185 pKa = 6.14KK186 pKa = 10.44RR187 pKa = 11.84KK188 pKa = 10.3EE189 pKa = 3.84KK190 pKa = 10.28DD191 pKa = 3.01QKK193 pKa = 11.64AFLATDD199 pKa = 3.82WNNDD203 pKa = 3.68DD204 pKa = 5.69LSPEE208 pKa = 4.01DD209 pKa = 3.7WDD211 pKa = 4.93DD212 pKa = 4.49LEE214 pKa = 4.4EE215 pKa = 4.25QAAHH219 pKa = 5.73YY220 pKa = 10.51HH221 pKa = 7.21DD222 pKa = 5.91DD223 pKa = 4.67DD224 pKa = 4.55EE225 pKa = 7.21LILPVKK231 pKa = 10.53RR232 pKa = 11.84KK233 pKa = 9.02VDD235 pKa = 3.44KK236 pKa = 10.82KK237 pKa = 11.02KK238 pKa = 10.47PLALRR243 pKa = 11.84RR244 pKa = 11.84KK245 pKa = 8.39PLPPVGFAGAMAEE258 pKa = 3.92ARR260 pKa = 11.84EE261 pKa = 4.19KK262 pKa = 11.38GDD264 pKa = 3.29LTFTFPVVFMGEE276 pKa = 4.1SDD278 pKa = 5.49DD279 pKa = 5.77DD280 pKa = 4.05DD281 pKa = 4.37TPVWEE286 pKa = 4.43PLPLKK291 pKa = 8.8TLKK294 pKa = 10.12EE295 pKa = 4.02LQSAVRR301 pKa = 11.84TMGPSAPYY309 pKa = 8.48TLQVVDD315 pKa = 4.68MVASQWLTPSDD326 pKa = 2.96WHH328 pKa = 4.57QTARR332 pKa = 11.84ATLSPGDD339 pKa = 3.64YY340 pKa = 10.37VLWRR344 pKa = 11.84TEE346 pKa = 3.81YY347 pKa = 10.48EE348 pKa = 4.28EE349 pKa = 4.34KK350 pKa = 10.82SKK352 pKa = 9.72EE353 pKa = 4.5TVQKK357 pKa = 9.56TAGKK361 pKa = 9.84RR362 pKa = 11.84KK363 pKa = 9.95GKK365 pKa = 10.54VSLDD369 pKa = 3.39MLLGTGQFLSPSSQIKK385 pKa = 10.11LSKK388 pKa = 10.67DD389 pKa = 3.14VLKK392 pKa = 10.93DD393 pKa = 3.24VTTNAVLAWRR403 pKa = 11.84AIPPPGVKK411 pKa = 8.73KK412 pKa = 9.08TVLAGLKK419 pKa = 9.73QGNEE423 pKa = 3.61EE424 pKa = 4.18SYY426 pKa = 9.06EE427 pKa = 4.17TFISRR432 pKa = 11.84LEE434 pKa = 3.95EE435 pKa = 3.46AVYY438 pKa = 11.09RR439 pKa = 11.84MMPRR443 pKa = 11.84GEE445 pKa = 4.33GSDD448 pKa = 3.43ILIKK452 pKa = 10.43QLAWEE457 pKa = 4.39NANSLCQDD465 pKa = 5.01LIRR468 pKa = 11.84PMRR471 pKa = 11.84KK472 pKa = 7.29TGTMQDD478 pKa = 3.89YY479 pKa = 10.81IRR481 pKa = 11.84ACLDD485 pKa = 3.11ASPAVVQGMAYY496 pKa = 9.19AAAMRR501 pKa = 11.84GQKK504 pKa = 10.41YY505 pKa = 7.44STFVKK510 pKa = 8.72QTYY513 pKa = 10.18GGGKK517 pKa = 9.54GGQGSEE523 pKa = 4.33GPVCFSCGKK532 pKa = 8.11TGHH535 pKa = 6.88IKK537 pKa = 10.2RR538 pKa = 11.84DD539 pKa = 3.61CKK541 pKa = 10.52EE542 pKa = 3.87EE543 pKa = 4.15KK544 pKa = 10.01GSKK547 pKa = 9.49RR548 pKa = 11.84APPGLCPRR556 pKa = 11.84CKK558 pKa = 10.06KK559 pKa = 10.1GYY561 pKa = 8.72HH562 pKa = 5.97WKK564 pKa = 10.34SEE566 pKa = 4.25CKK568 pKa = 10.4SKK570 pKa = 10.64FDD572 pKa = 4.67KK573 pKa = 11.01DD574 pKa = 4.08GNPLPPLEE582 pKa = 4.45TNAEE586 pKa = 3.94NSKK589 pKa = 10.69NLVKK593 pKa = 10.6GQSPSPTQKK602 pKa = 10.56GDD604 pKa = 2.96KK605 pKa = 10.95GKK607 pKa = 10.56DD608 pKa = 2.96SGLNPEE614 pKa = 4.78APPFTIHH621 pKa = 7.2DD622 pKa = 4.52LPRR625 pKa = 11.84GTPGSAGLDD634 pKa = 3.34LSSQKK639 pKa = 11.1DD640 pKa = 3.83LILSLEE646 pKa = 4.66DD647 pKa = 3.62GVSLVPTLVKK657 pKa = 9.96GTLPEE662 pKa = 4.4GTTGLIIGRR671 pKa = 11.84SSNYY675 pKa = 9.95KK676 pKa = 10.09KK677 pKa = 10.73GLEE680 pKa = 3.98VLPGVIDD687 pKa = 3.78SDD689 pKa = 3.88FQGEE693 pKa = 4.13IKK695 pKa = 11.16VMVKK699 pKa = 9.89AAKK702 pKa = 9.95NAVIIHH708 pKa = 5.75KK709 pKa = 9.37GEE711 pKa = 4.84RR712 pKa = 11.84IAQLLLLPYY721 pKa = 10.27LKK723 pKa = 10.6LPNPIIKK730 pKa = 9.82EE731 pKa = 4.0EE732 pKa = 4.09RR733 pKa = 11.84GSEE736 pKa = 4.39GFGSTSHH743 pKa = 5.12VHH745 pKa = 5.11WVQEE749 pKa = 4.27ISDD752 pKa = 4.49SRR754 pKa = 11.84PMLHH758 pKa = 6.63ISLNGRR764 pKa = 11.84RR765 pKa = 11.84FLGLLDD771 pKa = 3.56TGADD775 pKa = 3.57KK776 pKa = 10.91TCIAGRR782 pKa = 11.84DD783 pKa = 3.7WPANWPIHH791 pKa = 4.59QTEE794 pKa = 4.39SSLQGLGMACGVARR808 pKa = 11.84SSQPLRR814 pKa = 11.84WQHH817 pKa = 5.4EE818 pKa = 4.34DD819 pKa = 2.71KK820 pKa = 11.25SGIIHH825 pKa = 6.86PFVIPTLPFTLWGRR839 pKa = 11.84DD840 pKa = 2.7IMKK843 pKa = 9.72EE844 pKa = 3.58IKK846 pKa = 9.82VRR848 pKa = 11.84LMTDD852 pKa = 3.18SPDD855 pKa = 3.91DD856 pKa = 3.91SQDD859 pKa = 3.16LL860 pKa = 3.79
MM1 pKa = 7.24GVSGSKK7 pKa = 9.48GQKK10 pKa = 10.01LFVSVLQRR18 pKa = 11.84LLSEE22 pKa = 4.7RR23 pKa = 11.84GLHH26 pKa = 5.31VKK28 pKa = 10.05EE29 pKa = 4.39SSAIEE34 pKa = 4.43FYY36 pKa = 11.02QFLIKK41 pKa = 10.55VSPWFPEE48 pKa = 3.79EE49 pKa = 4.5GGLNLQDD56 pKa = 3.26WKK58 pKa = 11.05RR59 pKa = 11.84VGRR62 pKa = 11.84EE63 pKa = 3.7MKK65 pKa = 10.24KK66 pKa = 9.98YY67 pKa = 9.71AAEE70 pKa = 4.67HH71 pKa = 5.81GTDD74 pKa = 4.27SIPKK78 pKa = 7.84QAYY81 pKa = 7.84PIWLQLRR88 pKa = 11.84EE89 pKa = 4.17ILTEE93 pKa = 3.88QSDD96 pKa = 4.44LVLLSAEE103 pKa = 4.15AKK105 pKa = 10.18SVTEE109 pKa = 4.08EE110 pKa = 3.97EE111 pKa = 4.47LEE113 pKa = 4.17EE114 pKa = 4.46GLTGLLSASSQEE126 pKa = 3.85KK127 pKa = 9.34TYY129 pKa = 8.97GTRR132 pKa = 11.84GTAYY136 pKa = 10.45AEE138 pKa = 4.13IDD140 pKa = 3.78TEE142 pKa = 4.13VDD144 pKa = 3.01KK145 pKa = 11.4LSEE148 pKa = 4.53HH149 pKa = 7.04IYY151 pKa = 10.55DD152 pKa = 3.66EE153 pKa = 4.71PYY155 pKa = 10.62EE156 pKa = 4.21EE157 pKa = 5.61KK158 pKa = 10.79EE159 pKa = 4.01KK160 pKa = 10.96ADD162 pKa = 3.5KK163 pKa = 11.34NEE165 pKa = 4.22EE166 pKa = 3.82KK167 pKa = 11.02DD168 pKa = 3.3HH169 pKa = 5.17VRR171 pKa = 11.84KK172 pKa = 9.7VKK174 pKa = 10.63KK175 pKa = 9.75IVQRR179 pKa = 11.84KK180 pKa = 7.92EE181 pKa = 3.59NSEE184 pKa = 4.13HH185 pKa = 6.14KK186 pKa = 10.44RR187 pKa = 11.84KK188 pKa = 10.3EE189 pKa = 3.84KK190 pKa = 10.28DD191 pKa = 3.01QKK193 pKa = 11.64AFLATDD199 pKa = 3.82WNNDD203 pKa = 3.68DD204 pKa = 5.69LSPEE208 pKa = 4.01DD209 pKa = 3.7WDD211 pKa = 4.93DD212 pKa = 4.49LEE214 pKa = 4.4EE215 pKa = 4.25QAAHH219 pKa = 5.73YY220 pKa = 10.51HH221 pKa = 7.21DD222 pKa = 5.91DD223 pKa = 4.67DD224 pKa = 4.55EE225 pKa = 7.21LILPVKK231 pKa = 10.53RR232 pKa = 11.84KK233 pKa = 9.02VDD235 pKa = 3.44KK236 pKa = 10.82KK237 pKa = 11.02KK238 pKa = 10.47PLALRR243 pKa = 11.84RR244 pKa = 11.84KK245 pKa = 8.39PLPPVGFAGAMAEE258 pKa = 3.92ARR260 pKa = 11.84EE261 pKa = 4.19KK262 pKa = 11.38GDD264 pKa = 3.29LTFTFPVVFMGEE276 pKa = 4.1SDD278 pKa = 5.49DD279 pKa = 5.77DD280 pKa = 4.05DD281 pKa = 4.37TPVWEE286 pKa = 4.43PLPLKK291 pKa = 8.8TLKK294 pKa = 10.12EE295 pKa = 4.02LQSAVRR301 pKa = 11.84TMGPSAPYY309 pKa = 8.48TLQVVDD315 pKa = 4.68MVASQWLTPSDD326 pKa = 2.96WHH328 pKa = 4.57QTARR332 pKa = 11.84ATLSPGDD339 pKa = 3.64YY340 pKa = 10.37VLWRR344 pKa = 11.84TEE346 pKa = 3.81YY347 pKa = 10.48EE348 pKa = 4.28EE349 pKa = 4.34KK350 pKa = 10.82SKK352 pKa = 9.72EE353 pKa = 4.5TVQKK357 pKa = 9.56TAGKK361 pKa = 9.84RR362 pKa = 11.84KK363 pKa = 9.95GKK365 pKa = 10.54VSLDD369 pKa = 3.39MLLGTGQFLSPSSQIKK385 pKa = 10.11LSKK388 pKa = 10.67DD389 pKa = 3.14VLKK392 pKa = 10.93DD393 pKa = 3.24VTTNAVLAWRR403 pKa = 11.84AIPPPGVKK411 pKa = 8.73KK412 pKa = 9.08TVLAGLKK419 pKa = 9.73QGNEE423 pKa = 3.61EE424 pKa = 4.18SYY426 pKa = 9.06EE427 pKa = 4.17TFISRR432 pKa = 11.84LEE434 pKa = 3.95EE435 pKa = 3.46AVYY438 pKa = 11.09RR439 pKa = 11.84MMPRR443 pKa = 11.84GEE445 pKa = 4.33GSDD448 pKa = 3.43ILIKK452 pKa = 10.43QLAWEE457 pKa = 4.39NANSLCQDD465 pKa = 5.01LIRR468 pKa = 11.84PMRR471 pKa = 11.84KK472 pKa = 7.29TGTMQDD478 pKa = 3.89YY479 pKa = 10.81IRR481 pKa = 11.84ACLDD485 pKa = 3.11ASPAVVQGMAYY496 pKa = 9.19AAAMRR501 pKa = 11.84GQKK504 pKa = 10.41YY505 pKa = 7.44STFVKK510 pKa = 8.72QTYY513 pKa = 10.18GGGKK517 pKa = 9.54GGQGSEE523 pKa = 4.33GPVCFSCGKK532 pKa = 8.11TGHH535 pKa = 6.88IKK537 pKa = 10.2RR538 pKa = 11.84DD539 pKa = 3.61CKK541 pKa = 10.52EE542 pKa = 3.87EE543 pKa = 4.15KK544 pKa = 10.01GSKK547 pKa = 9.49RR548 pKa = 11.84APPGLCPRR556 pKa = 11.84CKK558 pKa = 10.06KK559 pKa = 10.1GYY561 pKa = 8.72HH562 pKa = 5.97WKK564 pKa = 10.34SEE566 pKa = 4.25CKK568 pKa = 10.4SKK570 pKa = 10.64FDD572 pKa = 4.67KK573 pKa = 11.01DD574 pKa = 4.08GNPLPPLEE582 pKa = 4.45TNAEE586 pKa = 3.94NSKK589 pKa = 10.69NLVKK593 pKa = 10.6GQSPSPTQKK602 pKa = 10.56GDD604 pKa = 2.96KK605 pKa = 10.95GKK607 pKa = 10.56DD608 pKa = 2.96SGLNPEE614 pKa = 4.78APPFTIHH621 pKa = 7.2DD622 pKa = 4.52LPRR625 pKa = 11.84GTPGSAGLDD634 pKa = 3.34LSSQKK639 pKa = 11.1DD640 pKa = 3.83LILSLEE646 pKa = 4.66DD647 pKa = 3.62GVSLVPTLVKK657 pKa = 9.96GTLPEE662 pKa = 4.4GTTGLIIGRR671 pKa = 11.84SSNYY675 pKa = 9.95KK676 pKa = 10.09KK677 pKa = 10.73GLEE680 pKa = 3.98VLPGVIDD687 pKa = 3.78SDD689 pKa = 3.88FQGEE693 pKa = 4.13IKK695 pKa = 11.16VMVKK699 pKa = 9.89AAKK702 pKa = 9.95NAVIIHH708 pKa = 5.75KK709 pKa = 9.37GEE711 pKa = 4.84RR712 pKa = 11.84IAQLLLLPYY721 pKa = 10.27LKK723 pKa = 10.6LPNPIIKK730 pKa = 9.82EE731 pKa = 4.0EE732 pKa = 4.09RR733 pKa = 11.84GSEE736 pKa = 4.39GFGSTSHH743 pKa = 5.12VHH745 pKa = 5.11WVQEE749 pKa = 4.27ISDD752 pKa = 4.49SRR754 pKa = 11.84PMLHH758 pKa = 6.63ISLNGRR764 pKa = 11.84RR765 pKa = 11.84FLGLLDD771 pKa = 3.56TGADD775 pKa = 3.57KK776 pKa = 10.91TCIAGRR782 pKa = 11.84DD783 pKa = 3.7WPANWPIHH791 pKa = 4.59QTEE794 pKa = 4.39SSLQGLGMACGVARR808 pKa = 11.84SSQPLRR814 pKa = 11.84WQHH817 pKa = 5.4EE818 pKa = 4.34DD819 pKa = 2.71KK820 pKa = 11.25SGIIHH825 pKa = 6.86PFVIPTLPFTLWGRR839 pKa = 11.84DD840 pKa = 2.7IMKK843 pKa = 9.72EE844 pKa = 3.58IKK846 pKa = 9.82VRR848 pKa = 11.84LMTDD852 pKa = 3.18SPDD855 pKa = 3.91DD856 pKa = 3.91SQDD859 pKa = 3.16LL860 pKa = 3.79
Molecular weight: 95.59 kDa
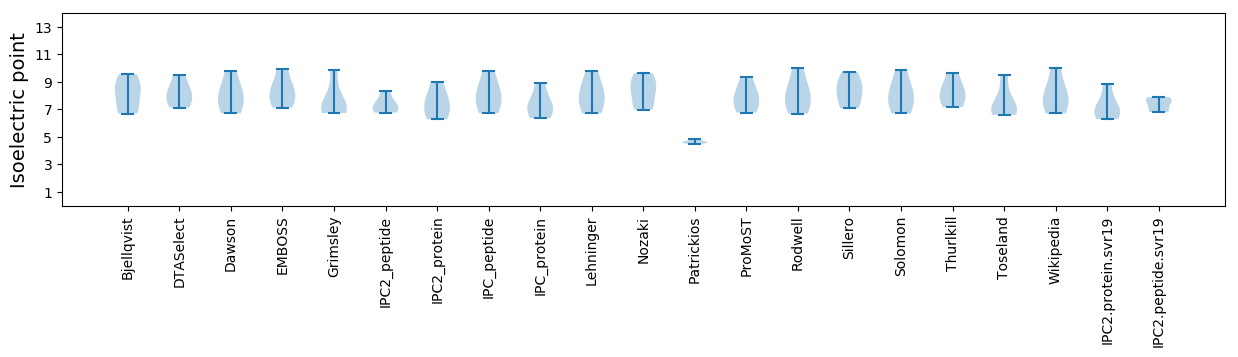
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P11283|POL_MMTVC Gag-Pro-Pol polyprotein OS=Mouse mammary tumor virus (strain C3H) OX=11759 GN=gag-pro-pol PE=1 SV=2
MM1 pKa = 7.81PRR3 pKa = 11.84LQQKK7 pKa = 8.42WLNSRR12 pKa = 11.84EE13 pKa = 4.26CPTPRR18 pKa = 11.84GEE20 pKa = 4.01AAKK23 pKa = 10.8GLFPTKK29 pKa = 10.09DD30 pKa = 3.35DD31 pKa = 4.04PSAHH35 pKa = 6.33KK36 pKa = 10.28RR37 pKa = 11.84VSPSDD42 pKa = 3.17KK43 pKa = 10.83DD44 pKa = 4.33IFILCCKK51 pKa = 10.39LGIALLCLGLLGEE64 pKa = 4.43VAVRR68 pKa = 11.84ARR70 pKa = 11.84RR71 pKa = 11.84ALTLDD76 pKa = 3.59SFNSSSVQDD85 pKa = 3.67YY86 pKa = 11.05NLNNSEE92 pKa = 4.01NSTFLLRR99 pKa = 11.84QGPQPTSSYY108 pKa = 10.03KK109 pKa = 9.43PHH111 pKa = 7.07RR112 pKa = 11.84FCPSEE117 pKa = 3.68IEE119 pKa = 3.97IRR121 pKa = 11.84MLAKK125 pKa = 10.53NYY127 pKa = 10.11IFTNKK132 pKa = 8.61TNPIGRR138 pKa = 11.84LLVTMLRR145 pKa = 11.84NEE147 pKa = 4.22SLSFSTIFTQIQKK160 pKa = 10.9LEE162 pKa = 3.86MGIEE166 pKa = 3.83NRR168 pKa = 11.84KK169 pKa = 9.38RR170 pKa = 11.84RR171 pKa = 11.84STSIEE176 pKa = 3.86EE177 pKa = 4.07QVQGLLTTGLEE188 pKa = 4.15VKK190 pKa = 10.08KK191 pKa = 10.57GKK193 pKa = 10.36KK194 pKa = 9.59SVFVKK199 pKa = 10.43IGDD202 pKa = 3.81RR203 pKa = 11.84WWQPGTYY210 pKa = 8.74RR211 pKa = 11.84GPYY214 pKa = 9.15IYY216 pKa = 10.27RR217 pKa = 11.84PTDD220 pKa = 3.06APLPYY225 pKa = 9.61TGRR228 pKa = 11.84YY229 pKa = 8.4DD230 pKa = 4.45LNWDD234 pKa = 3.05RR235 pKa = 11.84WVTVNGYY242 pKa = 9.47KK243 pKa = 10.11VLYY246 pKa = 9.63RR247 pKa = 11.84SLPFRR252 pKa = 11.84EE253 pKa = 3.71RR254 pKa = 11.84LARR257 pKa = 11.84ARR259 pKa = 11.84PPWCMLSQEE268 pKa = 5.0EE269 pKa = 4.47KK270 pKa = 11.09DD271 pKa = 3.96DD272 pKa = 3.95MKK274 pKa = 11.16QQVHH278 pKa = 6.95DD279 pKa = 4.61YY280 pKa = 9.71IYY282 pKa = 10.4LGTGMHH288 pKa = 6.45FWGKK292 pKa = 9.73IFHH295 pKa = 6.16TKK297 pKa = 9.89EE298 pKa = 3.66GTVAGLIEE306 pKa = 4.89HH307 pKa = 6.72YY308 pKa = 9.96SAKK311 pKa = 9.69TYY313 pKa = 10.37GMSYY317 pKa = 10.87YY318 pKa = 10.34EE319 pKa = 4.02
MM1 pKa = 7.81PRR3 pKa = 11.84LQQKK7 pKa = 8.42WLNSRR12 pKa = 11.84EE13 pKa = 4.26CPTPRR18 pKa = 11.84GEE20 pKa = 4.01AAKK23 pKa = 10.8GLFPTKK29 pKa = 10.09DD30 pKa = 3.35DD31 pKa = 4.04PSAHH35 pKa = 6.33KK36 pKa = 10.28RR37 pKa = 11.84VSPSDD42 pKa = 3.17KK43 pKa = 10.83DD44 pKa = 4.33IFILCCKK51 pKa = 10.39LGIALLCLGLLGEE64 pKa = 4.43VAVRR68 pKa = 11.84ARR70 pKa = 11.84RR71 pKa = 11.84ALTLDD76 pKa = 3.59SFNSSSVQDD85 pKa = 3.67YY86 pKa = 11.05NLNNSEE92 pKa = 4.01NSTFLLRR99 pKa = 11.84QGPQPTSSYY108 pKa = 10.03KK109 pKa = 9.43PHH111 pKa = 7.07RR112 pKa = 11.84FCPSEE117 pKa = 3.68IEE119 pKa = 3.97IRR121 pKa = 11.84MLAKK125 pKa = 10.53NYY127 pKa = 10.11IFTNKK132 pKa = 8.61TNPIGRR138 pKa = 11.84LLVTMLRR145 pKa = 11.84NEE147 pKa = 4.22SLSFSTIFTQIQKK160 pKa = 10.9LEE162 pKa = 3.86MGIEE166 pKa = 3.83NRR168 pKa = 11.84KK169 pKa = 9.38RR170 pKa = 11.84RR171 pKa = 11.84STSIEE176 pKa = 3.86EE177 pKa = 4.07QVQGLLTTGLEE188 pKa = 4.15VKK190 pKa = 10.08KK191 pKa = 10.57GKK193 pKa = 10.36KK194 pKa = 9.59SVFVKK199 pKa = 10.43IGDD202 pKa = 3.81RR203 pKa = 11.84WWQPGTYY210 pKa = 8.74RR211 pKa = 11.84GPYY214 pKa = 9.15IYY216 pKa = 10.27RR217 pKa = 11.84PTDD220 pKa = 3.06APLPYY225 pKa = 9.61TGRR228 pKa = 11.84YY229 pKa = 8.4DD230 pKa = 4.45LNWDD234 pKa = 3.05RR235 pKa = 11.84WVTVNGYY242 pKa = 9.47KK243 pKa = 10.11VLYY246 pKa = 9.63RR247 pKa = 11.84SLPFRR252 pKa = 11.84EE253 pKa = 3.71RR254 pKa = 11.84LARR257 pKa = 11.84ARR259 pKa = 11.84PPWCMLSQEE268 pKa = 5.0EE269 pKa = 4.47KK270 pKa = 11.09DD271 pKa = 3.96DD272 pKa = 3.95MKK274 pKa = 11.16QQVHH278 pKa = 6.95DD279 pKa = 4.61YY280 pKa = 9.71IYY282 pKa = 10.4LGTGMHH288 pKa = 6.45FWGKK292 pKa = 9.73IFHH295 pKa = 6.16TKK297 pKa = 9.89EE298 pKa = 3.66GTVAGLIEE306 pKa = 4.89HH307 pKa = 6.72YY308 pKa = 9.96SAKK311 pKa = 9.69TYY313 pKa = 10.37GMSYY317 pKa = 10.87YY318 pKa = 10.34EE319 pKa = 4.02
Molecular weight: 36.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4213 |
319 |
1755 |
842.6 |
94.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.29 ± 0.258 | 1.305 ± 0.11 |
5.72 ± 0.257 | 6.694 ± 0.636 |
3.181 ± 0.373 | 6.931 ± 0.426 |
2.421 ± 0.269 | 4.913 ± 0.467 |
8.189 ± 0.618 | 10.207 ± 0.279 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.875 ± 0.12 | 3.038 ± 0.384 |
6.361 ± 0.189 | 4.652 ± 0.306 |
4.937 ± 0.335 | 6.907 ± 0.21 |
5.839 ± 0.171 | 5.673 ± 0.148 |
2.089 ± 0.054 | 2.777 ± 0.288 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
