
Candidatus Scalindua japonica
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Candidatus Brocadiae; Candidatus Brocadiales; Candidatus Brocadiaceae; Candidatus Scalindua
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
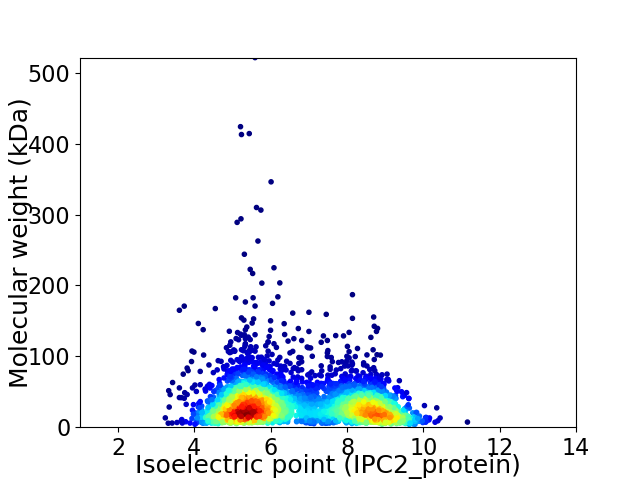
Virtual 2D-PAGE plot for 3991 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A286U0E9|A0A286U0E9_9BACT MerR-family transcriptional regulator OS=Candidatus Scalindua japonica OX=1284222 GN=SCALIN_C27_0009 PE=4 SV=1
MM1 pKa = 8.04SGMSKK6 pKa = 9.0TVVLVVARR14 pKa = 11.84CFVMVALLPCFMFSIASAQLNEE36 pKa = 4.15NCTVTVLNRR45 pKa = 11.84TVNVKK50 pKa = 10.26PDD52 pKa = 3.64GSWQLNNVPSNMGRR66 pKa = 11.84VRR68 pKa = 11.84VRR70 pKa = 11.84VTCVEE75 pKa = 4.16DD76 pKa = 3.63GKK78 pKa = 9.08TVSGQSEE85 pKa = 5.08YY86 pKa = 9.22VTIQTNGNISVEE98 pKa = 3.89EE99 pKa = 3.85MFFNNSYY106 pKa = 9.88EE107 pKa = 4.01QVPTSLTINSEE118 pKa = 3.88KK119 pKa = 10.72SVLSAIGEE127 pKa = 4.27TTRR130 pKa = 11.84ISVLAMYY137 pKa = 10.04PDD139 pKa = 4.87GSTKK143 pKa = 10.71DD144 pKa = 3.45VTLGGQGTAYY154 pKa = 9.27TVSNQAIATVNSNGLVTAISSGNIIISASNEE185 pKa = 3.94MVLSSIFLSVQLSGDD200 pKa = 3.49SDD202 pKa = 4.47GDD204 pKa = 3.79GLPDD208 pKa = 5.5DD209 pKa = 5.57FEE211 pKa = 4.74LTNGLNPNNPVDD223 pKa = 3.89SLEE226 pKa = 5.4DD227 pKa = 3.46MDD229 pKa = 6.07EE230 pKa = 4.75DD231 pKa = 4.19GLTNLEE237 pKa = 4.28EE238 pKa = 4.44FDD240 pKa = 5.45LGTGMNNADD249 pKa = 3.76TDD251 pKa = 3.88GDD253 pKa = 4.3GISDD257 pKa = 3.83GEE259 pKa = 4.33EE260 pKa = 3.57IVAGDD265 pKa = 4.2DD266 pKa = 4.22GYY268 pKa = 10.51VTNPLLGDD276 pKa = 3.69SDD278 pKa = 5.51GDD280 pKa = 4.42GIWDD284 pKa = 3.66GLEE287 pKa = 3.87VSSASDD293 pKa = 3.63PNDD296 pKa = 3.48AGSFDD301 pKa = 4.22LAATLDD307 pKa = 4.36FIKK310 pKa = 9.94VTPSNFALTFNTIMGEE326 pKa = 4.16STKK329 pKa = 10.58QLTVTGTFTDD339 pKa = 3.97GNTIDD344 pKa = 4.38LTPTGIGTNYY354 pKa = 10.0SSSDD358 pKa = 3.34LVVANFGVTAGLVYY372 pKa = 10.85AGTDD376 pKa = 3.33GTAVITVTNNGFSDD390 pKa = 3.61TAVFTVEE397 pKa = 4.43TFSPTPLSYY406 pKa = 11.42VDD408 pKa = 3.64ISGFANNVDD417 pKa = 3.32VGGDD421 pKa = 3.61YY422 pKa = 10.82AYY424 pKa = 10.93VAAGSTGLQVVDD436 pKa = 3.46VSDD439 pKa = 3.76RR440 pKa = 11.84VNPAVVASEE449 pKa = 4.18DD450 pKa = 3.73TLGNANDD457 pKa = 3.81VKK459 pKa = 11.25VDD461 pKa = 3.37GALAYY466 pKa = 9.73IADD469 pKa = 4.44GVSGLQIMDD478 pKa = 3.91ISNPLDD484 pKa = 3.74PVTVGSVDD492 pKa = 3.76TPGDD496 pKa = 3.72AYY498 pKa = 10.85DD499 pKa = 3.62VAVSGIYY506 pKa = 10.2AYY508 pKa = 10.81VADD511 pKa = 4.98GNAGLQIINVSVSNNPVIEE530 pKa = 4.35GTVVTSNIAKK540 pKa = 10.2GVDD543 pKa = 3.06IDD545 pKa = 4.17PEE547 pKa = 4.36GNLAVVADD555 pKa = 5.13GSAGVQVIDD564 pKa = 3.47ITTPSNAGIIGTVDD578 pKa = 3.1TNGNARR584 pKa = 11.84DD585 pKa = 3.67VVLRR589 pKa = 11.84NSIAYY594 pKa = 9.83VSDD597 pKa = 3.41YY598 pKa = 11.18TGGLKK603 pKa = 10.13AVNISNPTTPTIAKK617 pKa = 9.54SVEE620 pKa = 3.74GNYY623 pKa = 10.85LMDD626 pKa = 4.77AALFDD631 pKa = 4.28SLVFGADD638 pKa = 2.96VLSFNAVPAYY648 pKa = 10.3NISIPDD654 pKa = 3.81DD655 pKa = 3.74PVFTAIINFAQFRR668 pKa = 11.84DD669 pKa = 3.89DD670 pKa = 5.73DD671 pKa = 4.3GTGTDD676 pKa = 2.97ADD678 pKa = 3.64SRR680 pKa = 11.84YY681 pKa = 10.6LYY683 pKa = 9.55LTTSNGIMEE692 pKa = 4.94NGSSGDD698 pKa = 3.13TRR700 pKa = 11.84LYY702 pKa = 9.21IGQYY706 pKa = 10.95RR707 pKa = 11.84MVQDD711 pKa = 3.46IAGVPPTVSITSPLAGIDD729 pKa = 3.63VIEE732 pKa = 4.41GAKK735 pKa = 9.91IQVASDD741 pKa = 3.25AADD744 pKa = 3.82DD745 pKa = 4.01VQVASVSFIIDD756 pKa = 3.45GNVVFTDD763 pKa = 3.54TSAPYY768 pKa = 10.07EE769 pKa = 3.98FDD771 pKa = 3.32YY772 pKa = 10.73TVPLGISGVTISATALDD789 pKa = 4.14LANNAGASPDD799 pKa = 3.61LFVTVIPDD807 pKa = 3.81PLTTILGSLVDD818 pKa = 3.84INGGPMGGVTVTTVNNLSTTSLPDD842 pKa = 3.3GTFSISDD849 pKa = 3.57VPTVQGNIQVTASTNISGWPYY870 pKa = 10.27RR871 pKa = 11.84GNSPSIPAVPGGMTDD886 pKa = 2.91AGVIIIEE893 pKa = 4.11RR894 pKa = 11.84DD895 pKa = 3.67VVSITVDD902 pKa = 3.25TTIVTGNRR910 pKa = 11.84SMDD913 pKa = 4.13FKK915 pKa = 11.5ALTVDD920 pKa = 4.21GANLTVEE927 pKa = 4.8GPHH930 pKa = 5.59TFYY933 pKa = 11.09SITLLNNAKK942 pKa = 8.72LTHH945 pKa = 7.27PDD947 pKa = 2.95TTTTEE952 pKa = 4.32EE953 pKa = 4.12YY954 pKa = 10.81SLDD957 pKa = 3.25ITVSDD962 pKa = 4.2TLTIDD967 pKa = 3.1SDD969 pKa = 3.95STIDD973 pKa = 3.67VSGKK977 pKa = 10.78GYY979 pKa = 10.44LGGYY983 pKa = 8.6NGPGNYY989 pKa = 9.38SYY991 pKa = 11.51YY992 pKa = 10.98GRR994 pKa = 11.84TLGNTTTGGSYY1005 pKa = 10.2VRR1007 pKa = 11.84SGGSYY1012 pKa = 10.58GGLGGQTGSDD1022 pKa = 3.7SVNGVYY1028 pKa = 10.8GSFYY1032 pKa = 10.85NPNEE1036 pKa = 4.24LGSGGGGNSYY1046 pKa = 10.6TNGGYY1051 pKa = 10.33GGNGGGLVRR1060 pKa = 11.84ITAAEE1065 pKa = 3.77IVLNGNGSIKK1075 pKa = 10.93SNGGDD1080 pKa = 4.58GEE1082 pKa = 4.35DD1083 pKa = 4.05SDD1085 pKa = 4.55WNGGGSGGGIYY1096 pKa = 10.18INVGTIRR1103 pKa = 11.84GDD1105 pKa = 3.44GSGTIAANGGKK1116 pKa = 8.28ATRR1119 pKa = 11.84SYY1121 pKa = 11.44SSTNGGGGGRR1131 pKa = 11.84IAVYY1135 pKa = 10.54YY1136 pKa = 10.69DD1137 pKa = 4.78DD1138 pKa = 5.2IAGFDD1143 pKa = 3.62INSITAYY1150 pKa = 10.4GGLGSSNDD1158 pKa = 3.47GGAGTIYY1165 pKa = 10.74LKK1167 pKa = 10.94DD1168 pKa = 3.52VVNKK1172 pKa = 10.3KK1173 pKa = 9.12EE1174 pKa = 3.76QLIGNNSNNTGKK1186 pKa = 8.14PTPLVSEE1193 pKa = 5.07DD1194 pKa = 3.63GNGFTFDD1201 pKa = 4.42DD1202 pKa = 5.13LIIGGGSISIDD1213 pKa = 3.11KK1214 pKa = 11.02LIVNKK1219 pKa = 9.01LTLDD1223 pKa = 3.92AGDD1226 pKa = 3.54SCMGEE1231 pKa = 3.7LVVNNVVVGDD1241 pKa = 3.79GALIFAEE1248 pKa = 4.19VMTVVNNLTVDD1259 pKa = 3.37NGEE1262 pKa = 4.17LSVVDD1267 pKa = 4.68LSVGNNVTLQNNGILTHH1284 pKa = 6.56EE1285 pKa = 4.67AATMTQAYY1293 pKa = 9.7SSTIGVGGTLAIDD1306 pKa = 3.49SGSTIDD1312 pKa = 3.99VSCKK1316 pKa = 10.49GYY1318 pKa = 10.5LGGYY1322 pKa = 8.56NGSGNYY1328 pKa = 9.67SYY1330 pKa = 11.48YY1331 pKa = 10.98GRR1333 pKa = 11.84TLGNTTTGGSYY1344 pKa = 10.2VRR1346 pKa = 11.84SGGSYY1351 pKa = 10.58GGLGGQTGTDD1361 pKa = 3.38NVNGVYY1367 pKa = 10.75GSFYY1371 pKa = 10.85NPNEE1375 pKa = 4.24LGSGGGGNSYY1385 pKa = 10.6TNGGYY1390 pKa = 10.33GGNGGGLVRR1399 pKa = 11.84ITAAEE1404 pKa = 3.77IVLNGNGSIKK1414 pKa = 10.93SNGGDD1419 pKa = 4.58GEE1421 pKa = 4.35DD1422 pKa = 4.05SDD1424 pKa = 4.55WNGGGSGGGIYY1435 pKa = 10.18INVGTIRR1442 pKa = 11.84GDD1444 pKa = 3.44GSGTIAANGGKK1455 pKa = 8.28ATRR1458 pKa = 11.84SYY1460 pKa = 11.44SSTNGGGGGRR1470 pKa = 11.84IAVYY1474 pKa = 10.54YY1475 pKa = 10.69DD1476 pKa = 4.78DD1477 pKa = 5.2IAGFDD1482 pKa = 3.62INSITAYY1489 pKa = 10.4GGLGSSNDD1497 pKa = 3.47GGAGTIYY1504 pKa = 10.57LKK1506 pKa = 11.0ASIQTYY1512 pKa = 10.8GDD1514 pKa = 4.41LIVDD1518 pKa = 4.1NNGSTSLVYY1527 pKa = 9.12STPLVSVGTGLSSDD1541 pKa = 3.63LTANTLTDD1549 pKa = 3.55STRR1552 pKa = 11.84KK1553 pKa = 9.22WNVDD1557 pKa = 2.94GLIGIFLNPNTSQEE1571 pKa = 4.28TTPATVFRR1579 pKa = 11.84ILSNDD1584 pKa = 2.99VTSITVEE1591 pKa = 3.79NTLNNLTDD1599 pKa = 3.43VTISGNSYY1607 pKa = 10.01IGEE1610 pKa = 4.33HH1611 pKa = 6.17YY1612 pKa = 10.4FDD1614 pKa = 4.85NLTIINGARR1623 pKa = 11.84VEE1625 pKa = 4.22TLDD1628 pKa = 4.71RR1629 pKa = 11.84ISFSYY1634 pKa = 10.72ILITSGGEE1642 pKa = 3.81IQADD1646 pKa = 3.38NYY1648 pKa = 9.42YY1649 pKa = 10.61QIGKK1653 pKa = 10.19AEE1655 pKa = 4.1TSLKK1659 pKa = 10.88DD1660 pKa = 4.08KK1661 pKa = 10.29LAKK1664 pKa = 9.17YY1665 pKa = 9.25QKK1667 pKa = 10.42KK1668 pKa = 10.01
MM1 pKa = 8.04SGMSKK6 pKa = 9.0TVVLVVARR14 pKa = 11.84CFVMVALLPCFMFSIASAQLNEE36 pKa = 4.15NCTVTVLNRR45 pKa = 11.84TVNVKK50 pKa = 10.26PDD52 pKa = 3.64GSWQLNNVPSNMGRR66 pKa = 11.84VRR68 pKa = 11.84VRR70 pKa = 11.84VTCVEE75 pKa = 4.16DD76 pKa = 3.63GKK78 pKa = 9.08TVSGQSEE85 pKa = 5.08YY86 pKa = 9.22VTIQTNGNISVEE98 pKa = 3.89EE99 pKa = 3.85MFFNNSYY106 pKa = 9.88EE107 pKa = 4.01QVPTSLTINSEE118 pKa = 3.88KK119 pKa = 10.72SVLSAIGEE127 pKa = 4.27TTRR130 pKa = 11.84ISVLAMYY137 pKa = 10.04PDD139 pKa = 4.87GSTKK143 pKa = 10.71DD144 pKa = 3.45VTLGGQGTAYY154 pKa = 9.27TVSNQAIATVNSNGLVTAISSGNIIISASNEE185 pKa = 3.94MVLSSIFLSVQLSGDD200 pKa = 3.49SDD202 pKa = 4.47GDD204 pKa = 3.79GLPDD208 pKa = 5.5DD209 pKa = 5.57FEE211 pKa = 4.74LTNGLNPNNPVDD223 pKa = 3.89SLEE226 pKa = 5.4DD227 pKa = 3.46MDD229 pKa = 6.07EE230 pKa = 4.75DD231 pKa = 4.19GLTNLEE237 pKa = 4.28EE238 pKa = 4.44FDD240 pKa = 5.45LGTGMNNADD249 pKa = 3.76TDD251 pKa = 3.88GDD253 pKa = 4.3GISDD257 pKa = 3.83GEE259 pKa = 4.33EE260 pKa = 3.57IVAGDD265 pKa = 4.2DD266 pKa = 4.22GYY268 pKa = 10.51VTNPLLGDD276 pKa = 3.69SDD278 pKa = 5.51GDD280 pKa = 4.42GIWDD284 pKa = 3.66GLEE287 pKa = 3.87VSSASDD293 pKa = 3.63PNDD296 pKa = 3.48AGSFDD301 pKa = 4.22LAATLDD307 pKa = 4.36FIKK310 pKa = 9.94VTPSNFALTFNTIMGEE326 pKa = 4.16STKK329 pKa = 10.58QLTVTGTFTDD339 pKa = 3.97GNTIDD344 pKa = 4.38LTPTGIGTNYY354 pKa = 10.0SSSDD358 pKa = 3.34LVVANFGVTAGLVYY372 pKa = 10.85AGTDD376 pKa = 3.33GTAVITVTNNGFSDD390 pKa = 3.61TAVFTVEE397 pKa = 4.43TFSPTPLSYY406 pKa = 11.42VDD408 pKa = 3.64ISGFANNVDD417 pKa = 3.32VGGDD421 pKa = 3.61YY422 pKa = 10.82AYY424 pKa = 10.93VAAGSTGLQVVDD436 pKa = 3.46VSDD439 pKa = 3.76RR440 pKa = 11.84VNPAVVASEE449 pKa = 4.18DD450 pKa = 3.73TLGNANDD457 pKa = 3.81VKK459 pKa = 11.25VDD461 pKa = 3.37GALAYY466 pKa = 9.73IADD469 pKa = 4.44GVSGLQIMDD478 pKa = 3.91ISNPLDD484 pKa = 3.74PVTVGSVDD492 pKa = 3.76TPGDD496 pKa = 3.72AYY498 pKa = 10.85DD499 pKa = 3.62VAVSGIYY506 pKa = 10.2AYY508 pKa = 10.81VADD511 pKa = 4.98GNAGLQIINVSVSNNPVIEE530 pKa = 4.35GTVVTSNIAKK540 pKa = 10.2GVDD543 pKa = 3.06IDD545 pKa = 4.17PEE547 pKa = 4.36GNLAVVADD555 pKa = 5.13GSAGVQVIDD564 pKa = 3.47ITTPSNAGIIGTVDD578 pKa = 3.1TNGNARR584 pKa = 11.84DD585 pKa = 3.67VVLRR589 pKa = 11.84NSIAYY594 pKa = 9.83VSDD597 pKa = 3.41YY598 pKa = 11.18TGGLKK603 pKa = 10.13AVNISNPTTPTIAKK617 pKa = 9.54SVEE620 pKa = 3.74GNYY623 pKa = 10.85LMDD626 pKa = 4.77AALFDD631 pKa = 4.28SLVFGADD638 pKa = 2.96VLSFNAVPAYY648 pKa = 10.3NISIPDD654 pKa = 3.81DD655 pKa = 3.74PVFTAIINFAQFRR668 pKa = 11.84DD669 pKa = 3.89DD670 pKa = 5.73DD671 pKa = 4.3GTGTDD676 pKa = 2.97ADD678 pKa = 3.64SRR680 pKa = 11.84YY681 pKa = 10.6LYY683 pKa = 9.55LTTSNGIMEE692 pKa = 4.94NGSSGDD698 pKa = 3.13TRR700 pKa = 11.84LYY702 pKa = 9.21IGQYY706 pKa = 10.95RR707 pKa = 11.84MVQDD711 pKa = 3.46IAGVPPTVSITSPLAGIDD729 pKa = 3.63VIEE732 pKa = 4.41GAKK735 pKa = 9.91IQVASDD741 pKa = 3.25AADD744 pKa = 3.82DD745 pKa = 4.01VQVASVSFIIDD756 pKa = 3.45GNVVFTDD763 pKa = 3.54TSAPYY768 pKa = 10.07EE769 pKa = 3.98FDD771 pKa = 3.32YY772 pKa = 10.73TVPLGISGVTISATALDD789 pKa = 4.14LANNAGASPDD799 pKa = 3.61LFVTVIPDD807 pKa = 3.81PLTTILGSLVDD818 pKa = 3.84INGGPMGGVTVTTVNNLSTTSLPDD842 pKa = 3.3GTFSISDD849 pKa = 3.57VPTVQGNIQVTASTNISGWPYY870 pKa = 10.27RR871 pKa = 11.84GNSPSIPAVPGGMTDD886 pKa = 2.91AGVIIIEE893 pKa = 4.11RR894 pKa = 11.84DD895 pKa = 3.67VVSITVDD902 pKa = 3.25TTIVTGNRR910 pKa = 11.84SMDD913 pKa = 4.13FKK915 pKa = 11.5ALTVDD920 pKa = 4.21GANLTVEE927 pKa = 4.8GPHH930 pKa = 5.59TFYY933 pKa = 11.09SITLLNNAKK942 pKa = 8.72LTHH945 pKa = 7.27PDD947 pKa = 2.95TTTTEE952 pKa = 4.32EE953 pKa = 4.12YY954 pKa = 10.81SLDD957 pKa = 3.25ITVSDD962 pKa = 4.2TLTIDD967 pKa = 3.1SDD969 pKa = 3.95STIDD973 pKa = 3.67VSGKK977 pKa = 10.78GYY979 pKa = 10.44LGGYY983 pKa = 8.6NGPGNYY989 pKa = 9.38SYY991 pKa = 11.51YY992 pKa = 10.98GRR994 pKa = 11.84TLGNTTTGGSYY1005 pKa = 10.2VRR1007 pKa = 11.84SGGSYY1012 pKa = 10.58GGLGGQTGSDD1022 pKa = 3.7SVNGVYY1028 pKa = 10.8GSFYY1032 pKa = 10.85NPNEE1036 pKa = 4.24LGSGGGGNSYY1046 pKa = 10.6TNGGYY1051 pKa = 10.33GGNGGGLVRR1060 pKa = 11.84ITAAEE1065 pKa = 3.77IVLNGNGSIKK1075 pKa = 10.93SNGGDD1080 pKa = 4.58GEE1082 pKa = 4.35DD1083 pKa = 4.05SDD1085 pKa = 4.55WNGGGSGGGIYY1096 pKa = 10.18INVGTIRR1103 pKa = 11.84GDD1105 pKa = 3.44GSGTIAANGGKK1116 pKa = 8.28ATRR1119 pKa = 11.84SYY1121 pKa = 11.44SSTNGGGGGRR1131 pKa = 11.84IAVYY1135 pKa = 10.54YY1136 pKa = 10.69DD1137 pKa = 4.78DD1138 pKa = 5.2IAGFDD1143 pKa = 3.62INSITAYY1150 pKa = 10.4GGLGSSNDD1158 pKa = 3.47GGAGTIYY1165 pKa = 10.74LKK1167 pKa = 10.94DD1168 pKa = 3.52VVNKK1172 pKa = 10.3KK1173 pKa = 9.12EE1174 pKa = 3.76QLIGNNSNNTGKK1186 pKa = 8.14PTPLVSEE1193 pKa = 5.07DD1194 pKa = 3.63GNGFTFDD1201 pKa = 4.42DD1202 pKa = 5.13LIIGGGSISIDD1213 pKa = 3.11KK1214 pKa = 11.02LIVNKK1219 pKa = 9.01LTLDD1223 pKa = 3.92AGDD1226 pKa = 3.54SCMGEE1231 pKa = 3.7LVVNNVVVGDD1241 pKa = 3.79GALIFAEE1248 pKa = 4.19VMTVVNNLTVDD1259 pKa = 3.37NGEE1262 pKa = 4.17LSVVDD1267 pKa = 4.68LSVGNNVTLQNNGILTHH1284 pKa = 6.56EE1285 pKa = 4.67AATMTQAYY1293 pKa = 9.7SSTIGVGGTLAIDD1306 pKa = 3.49SGSTIDD1312 pKa = 3.99VSCKK1316 pKa = 10.49GYY1318 pKa = 10.5LGGYY1322 pKa = 8.56NGSGNYY1328 pKa = 9.67SYY1330 pKa = 11.48YY1331 pKa = 10.98GRR1333 pKa = 11.84TLGNTTTGGSYY1344 pKa = 10.2VRR1346 pKa = 11.84SGGSYY1351 pKa = 10.58GGLGGQTGTDD1361 pKa = 3.38NVNGVYY1367 pKa = 10.75GSFYY1371 pKa = 10.85NPNEE1375 pKa = 4.24LGSGGGGNSYY1385 pKa = 10.6TNGGYY1390 pKa = 10.33GGNGGGLVRR1399 pKa = 11.84ITAAEE1404 pKa = 3.77IVLNGNGSIKK1414 pKa = 10.93SNGGDD1419 pKa = 4.58GEE1421 pKa = 4.35DD1422 pKa = 4.05SDD1424 pKa = 4.55WNGGGSGGGIYY1435 pKa = 10.18INVGTIRR1442 pKa = 11.84GDD1444 pKa = 3.44GSGTIAANGGKK1455 pKa = 8.28ATRR1458 pKa = 11.84SYY1460 pKa = 11.44SSTNGGGGGRR1470 pKa = 11.84IAVYY1474 pKa = 10.54YY1475 pKa = 10.69DD1476 pKa = 4.78DD1477 pKa = 5.2IAGFDD1482 pKa = 3.62INSITAYY1489 pKa = 10.4GGLGSSNDD1497 pKa = 3.47GGAGTIYY1504 pKa = 10.57LKK1506 pKa = 11.0ASIQTYY1512 pKa = 10.8GDD1514 pKa = 4.41LIVDD1518 pKa = 4.1NNGSTSLVYY1527 pKa = 9.12STPLVSVGTGLSSDD1541 pKa = 3.63LTANTLTDD1549 pKa = 3.55STRR1552 pKa = 11.84KK1553 pKa = 9.22WNVDD1557 pKa = 2.94GLIGIFLNPNTSQEE1571 pKa = 4.28TTPATVFRR1579 pKa = 11.84ILSNDD1584 pKa = 2.99VTSITVEE1591 pKa = 3.79NTLNNLTDD1599 pKa = 3.43VTISGNSYY1607 pKa = 10.01IGEE1610 pKa = 4.33HH1611 pKa = 6.17YY1612 pKa = 10.4FDD1614 pKa = 4.85NLTIINGARR1623 pKa = 11.84VEE1625 pKa = 4.22TLDD1628 pKa = 4.71RR1629 pKa = 11.84ISFSYY1634 pKa = 10.72ILITSGGEE1642 pKa = 3.81IQADD1646 pKa = 3.38NYY1648 pKa = 9.42YY1649 pKa = 10.61QIGKK1653 pKa = 10.19AEE1655 pKa = 4.1TSLKK1659 pKa = 10.88DD1660 pKa = 4.08KK1661 pKa = 10.29LAKK1664 pKa = 9.17YY1665 pKa = 9.25QKK1667 pKa = 10.42KK1668 pKa = 10.01
Molecular weight: 170.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A286U2R1|A0A286U2R1_9BACT Formylglycine-generating sulfatase-like enzyme OS=Candidatus Scalindua japonica OX=1284222 GN=SCALIN_C31_0051 PE=4 SV=1
MM1 pKa = 7.84PKK3 pKa = 10.57LKK5 pKa = 8.67THH7 pKa = 6.6KK8 pKa = 10.66GLTKK12 pKa = 10.07RR13 pKa = 11.84VKK15 pKa = 10.11VSPNGKK21 pKa = 8.42IKK23 pKa = 10.24RR24 pKa = 11.84RR25 pKa = 11.84KK26 pKa = 9.38AFAGHH31 pKa = 6.93LMSGKK36 pKa = 8.3TGNRR40 pKa = 11.84KK41 pKa = 9.04RR42 pKa = 11.84ALKK45 pKa = 9.31RR46 pKa = 11.84TSLVSKK52 pKa = 10.16GFIRR56 pKa = 11.84PLMRR60 pKa = 11.84ALGKK64 pKa = 10.33AA65 pKa = 3.3
MM1 pKa = 7.84PKK3 pKa = 10.57LKK5 pKa = 8.67THH7 pKa = 6.6KK8 pKa = 10.66GLTKK12 pKa = 10.07RR13 pKa = 11.84VKK15 pKa = 10.11VSPNGKK21 pKa = 8.42IKK23 pKa = 10.24RR24 pKa = 11.84RR25 pKa = 11.84KK26 pKa = 9.38AFAGHH31 pKa = 6.93LMSGKK36 pKa = 8.3TGNRR40 pKa = 11.84KK41 pKa = 9.04RR42 pKa = 11.84ALKK45 pKa = 9.31RR46 pKa = 11.84TSLVSKK52 pKa = 10.16GFIRR56 pKa = 11.84PLMRR60 pKa = 11.84ALGKK64 pKa = 10.33AA65 pKa = 3.3
Molecular weight: 7.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1248076 |
33 |
4670 |
312.7 |
35.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.136 ± 0.033 | 1.265 ± 0.016 |
5.77 ± 0.029 | 7.127 ± 0.032 |
4.345 ± 0.027 | 6.833 ± 0.041 |
2.02 ± 0.019 | 8.21 ± 0.039 |
7.826 ± 0.041 | 9.081 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.395 ± 0.017 | 5.012 ± 0.027 |
3.588 ± 0.022 | 2.955 ± 0.019 |
4.618 ± 0.03 | 6.49 ± 0.031 |
5.247 ± 0.032 | 6.604 ± 0.033 |
1.025 ± 0.013 | 3.455 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
