
Hortaea thailandica
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetidae; Mycosphaerellales; Teratosphaeriaceae; Hortaea
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
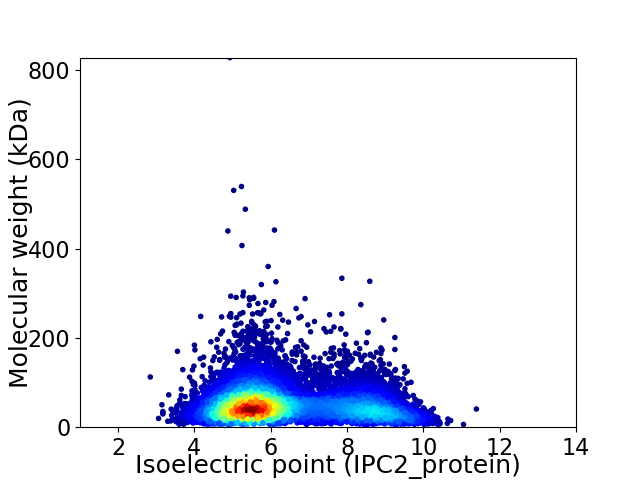
Virtual 2D-PAGE plot for 8771 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
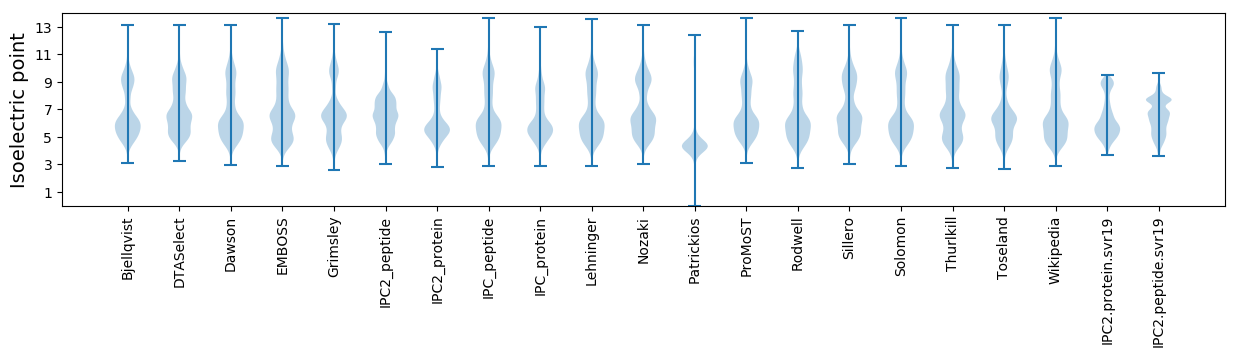
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4U0UA26|A0A4U0UA26_9PEZI Uncharacterized protein OS=Hortaea thailandica OX=706561 GN=B0A50_01365 PE=4 SV=1
MM1 pKa = 7.13FASFLLLSGLAALAAAQSTVLTFTHH26 pKa = 6.16VPNPVTDD33 pKa = 4.81GEE35 pKa = 4.3AQALTFSTDD44 pKa = 3.33DD45 pKa = 3.19ASNPVTITLLQGAANNLQTVSVLTTNAVNGQYY77 pKa = 10.6VWTPSADD84 pKa = 3.7LPNGADD90 pKa = 3.4YY91 pKa = 11.29ALEE94 pKa = 4.03IQQAGEE100 pKa = 4.05TNYY103 pKa = 10.24FGPFTVQGAGSVSSAMSSASSMPASSSMISASSMAPTSSMAPTSYY148 pKa = 9.89MAPTSSMLPASSMAPVSTAPSMSMNSTAATNTPMPVGTMGTAIMMNGTIYY198 pKa = 10.5PSGTAMPRR206 pKa = 11.84NTTMSMATLTPTTAMGGSPSTNSAAATGGSSAAPASSSMATNGATGTIAALSTTFLDD263 pKa = 5.4LYY265 pKa = 10.97NKK267 pKa = 8.43TSNMSPRR274 pKa = 11.84SLLLALVSAPLLAFAQSANPFSIPDD299 pKa = 3.66AGLSATGGQPLEE311 pKa = 4.77LMWNPTTDD319 pKa = 3.24GTVTLVLRR327 pKa = 11.84SGNADD332 pKa = 3.16NLAEE336 pKa = 4.2GTVIASNIDD345 pKa = 3.26NSGSYY350 pKa = 8.25TWTPSDD356 pKa = 3.41SLTRR360 pKa = 11.84GSDD363 pKa = 3.41YY364 pKa = 10.83TIEE367 pKa = 4.37IVDD370 pKa = 5.01DD371 pKa = 4.65DD372 pKa = 4.87DD373 pKa = 4.67VSNTNFTPYY382 pKa = 10.12FVLDD386 pKa = 3.46TTTTVAQSTSQVTLGAPTEE405 pKa = 4.37TPDD408 pKa = 4.28KK409 pKa = 11.0SSASPTGAATSVLDD423 pKa = 3.56ATMDD427 pKa = 3.64TSSAAATTMSSMSADD442 pKa = 3.46SSSAPASSSMASSSPTSMDD461 pKa = 3.14AAATTSAPAGKK472 pKa = 8.13MTMSAGNMTMGAGAPSAGSSTNSSASPSSSVEE504 pKa = 3.74PFIGGAASLKK514 pKa = 10.42VSAALGAFGLVALCAMVLL532 pKa = 3.58
MM1 pKa = 7.13FASFLLLSGLAALAAAQSTVLTFTHH26 pKa = 6.16VPNPVTDD33 pKa = 4.81GEE35 pKa = 4.3AQALTFSTDD44 pKa = 3.33DD45 pKa = 3.19ASNPVTITLLQGAANNLQTVSVLTTNAVNGQYY77 pKa = 10.6VWTPSADD84 pKa = 3.7LPNGADD90 pKa = 3.4YY91 pKa = 11.29ALEE94 pKa = 4.03IQQAGEE100 pKa = 4.05TNYY103 pKa = 10.24FGPFTVQGAGSVSSAMSSASSMPASSSMISASSMAPTSSMAPTSYY148 pKa = 9.89MAPTSSMLPASSMAPVSTAPSMSMNSTAATNTPMPVGTMGTAIMMNGTIYY198 pKa = 10.5PSGTAMPRR206 pKa = 11.84NTTMSMATLTPTTAMGGSPSTNSAAATGGSSAAPASSSMATNGATGTIAALSTTFLDD263 pKa = 5.4LYY265 pKa = 10.97NKK267 pKa = 8.43TSNMSPRR274 pKa = 11.84SLLLALVSAPLLAFAQSANPFSIPDD299 pKa = 3.66AGLSATGGQPLEE311 pKa = 4.77LMWNPTTDD319 pKa = 3.24GTVTLVLRR327 pKa = 11.84SGNADD332 pKa = 3.16NLAEE336 pKa = 4.2GTVIASNIDD345 pKa = 3.26NSGSYY350 pKa = 8.25TWTPSDD356 pKa = 3.41SLTRR360 pKa = 11.84GSDD363 pKa = 3.41YY364 pKa = 10.83TIEE367 pKa = 4.37IVDD370 pKa = 5.01DD371 pKa = 4.65DD372 pKa = 4.87DD373 pKa = 4.67VSNTNFTPYY382 pKa = 10.12FVLDD386 pKa = 3.46TTTTVAQSTSQVTLGAPTEE405 pKa = 4.37TPDD408 pKa = 4.28KK409 pKa = 11.0SSASPTGAATSVLDD423 pKa = 3.56ATMDD427 pKa = 3.64TSSAAATTMSSMSADD442 pKa = 3.46SSSAPASSSMASSSPTSMDD461 pKa = 3.14AAATTSAPAGKK472 pKa = 8.13MTMSAGNMTMGAGAPSAGSSTNSSASPSSSVEE504 pKa = 3.74PFIGGAASLKK514 pKa = 10.42VSAALGAFGLVALCAMVLL532 pKa = 3.58
Molecular weight: 52.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4U0TX96|A0A4U0TX96_9PEZI Carboxylic ester hydrolase OS=Hortaea thailandica OX=706561 GN=B0A50_05291 PE=3 SV=1
MM1 pKa = 7.35AAVVTPSSQPAPVARR16 pKa = 11.84APAARR21 pKa = 11.84VPAARR26 pKa = 11.84APVAKK31 pKa = 10.57APVAPVARR39 pKa = 11.84PVAATRR45 pKa = 11.84TAAAPVASSVKK56 pKa = 9.39PVTPARR62 pKa = 11.84PAVKK66 pKa = 10.14SPSTVPVAGRR76 pKa = 11.84PTGTSAPVTTGAATRR91 pKa = 11.84APARR95 pKa = 11.84SAAPVGVSGAAAVTPRR111 pKa = 11.84TTARR115 pKa = 11.84TTAATPAVANPVVPGRR131 pKa = 11.84RR132 pKa = 11.84AVGTTAATPVASRR145 pKa = 11.84TAAVTGTTAATRR157 pKa = 11.84TAVPATGAVPRR168 pKa = 11.84TAAPTTVGAAGVRR181 pKa = 11.84SAPVTTNGVRR191 pKa = 11.84TARR194 pKa = 11.84AAPATTAAVTPAGTTVARR212 pKa = 11.84PRR214 pKa = 11.84TATTGAATRR223 pKa = 11.84SGVVGAGRR231 pKa = 11.84KK232 pKa = 8.07VVGGGVAQPVSGNVAPVNNTIAGVRR257 pKa = 11.84QPVAGPGAVNGVVGNSVNTVGGPGRR282 pKa = 11.84AAIGSTAPAARR293 pKa = 11.84TVTPATSSVASTPVGSAAVTPAATRR318 pKa = 11.84AATPAVSGVAGGVAGAARR336 pKa = 11.84TVGGAVSSVGGAVPGVAGAARR357 pKa = 11.84PVGGAVSSVGGAVPGVAGAARR378 pKa = 11.84PVGGAVGGVGGAVADD393 pKa = 4.3PVTGAASTVGGAVPAAPTSAAAVPGVAGASGVVGNAVSGVGSTVAGVGRR442 pKa = 11.84SLGGLASGVVGG453 pKa = 4.44
MM1 pKa = 7.35AAVVTPSSQPAPVARR16 pKa = 11.84APAARR21 pKa = 11.84VPAARR26 pKa = 11.84APVAKK31 pKa = 10.57APVAPVARR39 pKa = 11.84PVAATRR45 pKa = 11.84TAAAPVASSVKK56 pKa = 9.39PVTPARR62 pKa = 11.84PAVKK66 pKa = 10.14SPSTVPVAGRR76 pKa = 11.84PTGTSAPVTTGAATRR91 pKa = 11.84APARR95 pKa = 11.84SAAPVGVSGAAAVTPRR111 pKa = 11.84TTARR115 pKa = 11.84TTAATPAVANPVVPGRR131 pKa = 11.84RR132 pKa = 11.84AVGTTAATPVASRR145 pKa = 11.84TAAVTGTTAATRR157 pKa = 11.84TAVPATGAVPRR168 pKa = 11.84TAAPTTVGAAGVRR181 pKa = 11.84SAPVTTNGVRR191 pKa = 11.84TARR194 pKa = 11.84AAPATTAAVTPAGTTVARR212 pKa = 11.84PRR214 pKa = 11.84TATTGAATRR223 pKa = 11.84SGVVGAGRR231 pKa = 11.84KK232 pKa = 8.07VVGGGVAQPVSGNVAPVNNTIAGVRR257 pKa = 11.84QPVAGPGAVNGVVGNSVNTVGGPGRR282 pKa = 11.84AAIGSTAPAARR293 pKa = 11.84TVTPATSSVASTPVGSAAVTPAATRR318 pKa = 11.84AATPAVSGVAGGVAGAARR336 pKa = 11.84TVGGAVSSVGGAVPGVAGAARR357 pKa = 11.84PVGGAVSSVGGAVPGVAGAARR378 pKa = 11.84PVGGAVGGVGGAVADD393 pKa = 4.3PVTGAASTVGGAVPAAPTSAAAVPGVAGASGVVGNAVSGVGSTVAGVGRR442 pKa = 11.84SLGGLASGVVGG453 pKa = 4.44
Molecular weight: 40.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4524954 |
51 |
7619 |
515.9 |
56.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.491 ± 0.022 | 1.095 ± 0.008 |
5.753 ± 0.019 | 6.429 ± 0.025 |
3.399 ± 0.017 | 7.442 ± 0.025 |
2.403 ± 0.01 | 4.11 ± 0.018 |
4.687 ± 0.024 | 8.558 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.226 ± 0.01 | 3.402 ± 0.015 |
6.191 ± 0.031 | 4.402 ± 0.022 |
6.315 ± 0.023 | 7.984 ± 0.034 |
6.059 ± 0.02 | 6.018 ± 0.018 |
1.372 ± 0.01 | 2.664 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
