
Criblamydia sequanensis CRIB-18
Taxonomy: cellular organisms; Bacteria; PVC group; Chlamydiae; Chlamydiia; Parachlamydiales; Criblamydiaceae; Criblamydia; Criblamydia sequanensis
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
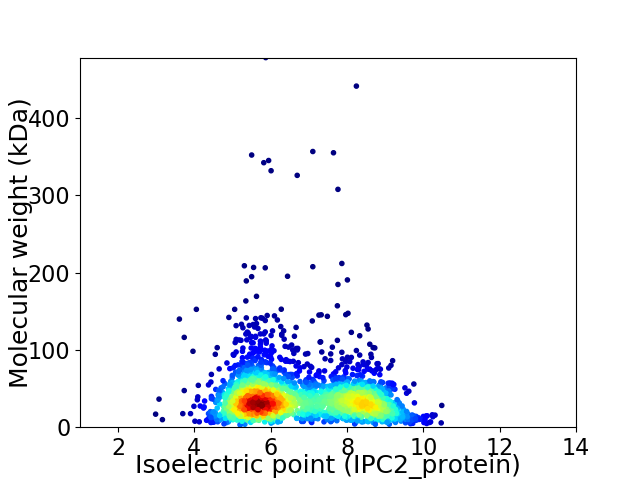
Virtual 2D-PAGE plot for 2418 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A090D0Q2|A0A090D0Q2_9BACT Putative secreted protein OS=Criblamydia sequanensis CRIB-18 OX=1437425 GN=CSEC_0314 PE=4 SV=1
MM1 pKa = 7.71KK2 pKa = 10.68LNILPLEE9 pKa = 4.0EE10 pKa = 5.59RR11 pKa = 11.84IVLDD15 pKa = 3.3AAAAVVIAAPPTATDD30 pKa = 3.24SGDD33 pKa = 3.57AATHH37 pKa = 6.9DD38 pKa = 4.14SSQATDD44 pKa = 3.21SSQEE48 pKa = 3.85SGPAQEE54 pKa = 4.79SGVNVLVISSAVQDD68 pKa = 3.9PQSLQNAAKK77 pKa = 10.44DD78 pKa = 3.73GVKK81 pKa = 9.57TVFYY85 pKa = 10.22DD86 pKa = 3.68ANSTSLTEE94 pKa = 4.31LSQKK98 pKa = 9.46IANALNGQKK107 pKa = 10.33ADD109 pKa = 4.02SIAFAAEE116 pKa = 4.06GNDD119 pKa = 3.27NVLMLNTDD127 pKa = 2.87IMVSDD132 pKa = 5.21NIQSNIQNHH141 pKa = 7.06DD142 pKa = 2.79ISGFWKK148 pKa = 9.62TIGGLLNNGGRR159 pKa = 11.84IDD161 pKa = 5.72LLACDD166 pKa = 3.91VAGDD170 pKa = 3.79SGGQSLVAQIDD181 pKa = 3.63QLIDD185 pKa = 3.38RR186 pKa = 11.84PQDD189 pKa = 3.21GFFATVAASTDD200 pKa = 3.59LTGNAAGGDD209 pKa = 3.4WLLEE213 pKa = 4.07VGNIDD218 pKa = 4.1AKK220 pKa = 10.75SIYY223 pKa = 10.6FNDD226 pKa = 5.03SINNWNHH233 pKa = 4.62NMATAIEE240 pKa = 4.25YY241 pKa = 10.94GLIAALIGALLQPRR255 pKa = 11.84EE256 pKa = 4.17TLEE259 pKa = 4.11DD260 pKa = 3.34TSLSFSYY267 pKa = 10.65ASSGTVFANPDD278 pKa = 3.26FSFTLLTNSINGNTTVTEE296 pKa = 4.66DD297 pKa = 3.63GNVTYY302 pKa = 9.98TPNADD307 pKa = 3.52YY308 pKa = 11.2NNPGFTLGGTINDD321 pKa = 3.7GVSFDD326 pKa = 3.56SATIRR331 pKa = 11.84IVDD334 pKa = 4.24TNPDD338 pKa = 3.52DD339 pKa = 4.55PNSVFAINLPLYY351 pKa = 10.49LRR353 pKa = 11.84VVSVNDD359 pKa = 3.61APIAQNDD366 pKa = 4.05SFSIGQGNEE375 pKa = 3.49LTGNVALNDD384 pKa = 3.58SDD386 pKa = 4.16FQNGAPSEE394 pKa = 4.38NNTPLTVQLVSGPAHH409 pKa = 6.16AASFTLNADD418 pKa = 3.3GTFSYY423 pKa = 10.62TSIGSYY429 pKa = 10.24SGADD433 pKa = 2.95SFTYY437 pKa = 10.5SITDD441 pKa = 3.29SLGGVSEE448 pKa = 4.72ANVALSVISNAPPAVSDD465 pKa = 3.5NYY467 pKa = 10.29IINEE471 pKa = 3.95DD472 pKa = 3.39TTLNGNVTDD481 pKa = 4.34NDD483 pKa = 4.2TFNLGANQTIQLVSGPLVAANFALNSDD510 pKa = 4.01GSFSYY515 pKa = 10.59TPLANLVGVDD525 pKa = 3.28TFSYY529 pKa = 10.28VIKK532 pKa = 8.94TVSGAGITFSNTATVNIAVLPVNDD556 pKa = 4.17NPVAINDD563 pKa = 3.78NYY565 pKa = 10.94NVNEE569 pKa = 4.79DD570 pKa = 3.17GSLAANVLNNDD581 pKa = 2.98TDD583 pKa = 3.68IEE585 pKa = 4.37NNRR588 pKa = 11.84PFSAQLVAGPAHH600 pKa = 6.62AGSFTLNADD609 pKa = 3.43GTFSYY614 pKa = 10.69IPTADD619 pKa = 3.63YY620 pKa = 11.14NGSDD624 pKa = 3.11SFTYY628 pKa = 9.45VTVDD632 pKa = 3.05SLGAISNLATVSIAVASINDD652 pKa = 3.53APVAQDD658 pKa = 2.72VSYY661 pKa = 9.18NTNEE665 pKa = 4.02DD666 pKa = 3.75TPLTGILGVGSDD678 pKa = 4.2SQNGAPNEE686 pKa = 4.49SNTPLTAQLIGGPAHH701 pKa = 7.27ASAFTLNADD710 pKa = 3.67GSFSYY715 pKa = 10.85NPQADD720 pKa = 3.96FNGTDD725 pKa = 3.01SFTYY729 pKa = 10.27AIVDD733 pKa = 3.62SLGAISNTATVSITIDD749 pKa = 3.59SVNDD753 pKa = 3.36APIAQNDD760 pKa = 3.69AFQGNEE766 pKa = 3.96DD767 pKa = 3.47GSINGSVLLNDD778 pKa = 4.47SDD780 pKa = 4.27NQNGAPNEE788 pKa = 4.2NNSPLSAQLVTGPAHH803 pKa = 6.72AGSFTLNADD812 pKa = 3.38GTFTYY817 pKa = 10.23IPSADD822 pKa = 3.73YY823 pKa = 11.04NGSDD827 pKa = 2.98SFTYY831 pKa = 9.37VTLDD835 pKa = 3.29SLGGISNLATVSITINSVNDD855 pKa = 3.38APIAQNDD862 pKa = 4.16SYY864 pKa = 11.51QVNEE868 pKa = 4.2DD869 pKa = 3.18NGLNGNVALNDD880 pKa = 3.64SDD882 pKa = 4.23SQNGAPSEE890 pKa = 4.28NNGPLSAQLVTGPAHH905 pKa = 6.72AGSFTLNADD914 pKa = 3.43GTFSYY919 pKa = 10.62IPAANYY925 pKa = 10.17NGADD929 pKa = 3.15SFTYY933 pKa = 10.57VIVDD937 pKa = 3.55SLGATSNIATVAITINSVNDD957 pKa = 3.5APIVQNDD964 pKa = 3.61SFNIPGNLPYY974 pKa = 10.58SGNVLTNDD982 pKa = 3.68SDD984 pKa = 4.25NQGGAPNEE992 pKa = 4.0NNIPLSAQLVNGPAHH1007 pKa = 5.84ATSFILNADD1016 pKa = 3.64GTFSYY1021 pKa = 10.88QPANGFVGSDD1031 pKa = 2.88TFTYY1035 pKa = 10.4RR1036 pKa = 11.84VTDD1039 pKa = 3.64SLGGTSTIATVTITVAQAASANPILEE1065 pKa = 4.19LKK1067 pKa = 11.02GNINKK1072 pKa = 10.0NYY1074 pKa = 9.34IEE1076 pKa = 4.19NSGPVILSNNVKK1088 pKa = 8.65ITDD1091 pKa = 3.7SDD1093 pKa = 3.7SSNFDD1098 pKa = 3.08GGKK1101 pKa = 8.46LTVQIDD1107 pKa = 3.69QNGQSSDD1114 pKa = 3.06ILSIDD1119 pKa = 3.22NSTKK1123 pKa = 10.54VKK1125 pKa = 10.12TNANQDD1131 pKa = 2.6IYY1133 pKa = 11.73YY1134 pKa = 10.47NGLLVGHH1141 pKa = 5.5YY1142 pKa = 9.29TGGSGLASLEE1152 pKa = 3.74ITFNANATANVVSEE1166 pKa = 4.12VAEE1169 pKa = 4.42RR1170 pKa = 11.84IAYY1173 pKa = 8.97KK1174 pKa = 10.56NSSEE1178 pKa = 4.22NPSTADD1184 pKa = 3.33RR1185 pKa = 11.84SIRR1188 pKa = 11.84YY1189 pKa = 8.39QLSDD1193 pKa = 3.4GDD1195 pKa = 4.23SGISEE1200 pKa = 4.21AQYY1203 pKa = 10.56KK1204 pKa = 8.77IVRR1207 pKa = 11.84VFSVNDD1213 pKa = 3.54APDD1216 pKa = 3.06ISIADD1221 pKa = 3.44NTLTVNRR1228 pKa = 11.84GSITNIGALSGIQISDD1244 pKa = 3.19VDD1246 pKa = 3.69ASAGDD1251 pKa = 3.61RR1252 pKa = 11.84VRR1254 pKa = 11.84VVLKK1258 pKa = 10.84VLVGYY1263 pKa = 11.05LDD1265 pKa = 3.68FTDD1268 pKa = 5.09DD1269 pKa = 5.06LNDD1272 pKa = 3.36LTSIIFKK1279 pKa = 10.44RR1280 pKa = 11.84NPGDD1284 pKa = 3.49GLGSLDD1290 pKa = 3.84FTGTLADD1297 pKa = 3.4VNAALEE1303 pKa = 4.06RR1304 pKa = 11.84LTYY1307 pKa = 10.3RR1308 pKa = 11.84SRR1310 pKa = 11.84TNYY1313 pKa = 9.64IGLDD1317 pKa = 3.36ALTVGVSDD1325 pKa = 4.21MGSSGTFAIPRR1336 pKa = 11.84IDD1338 pKa = 4.99ADD1340 pKa = 3.53WLLLKK1345 pKa = 10.3IVV1347 pKa = 3.84
MM1 pKa = 7.71KK2 pKa = 10.68LNILPLEE9 pKa = 4.0EE10 pKa = 5.59RR11 pKa = 11.84IVLDD15 pKa = 3.3AAAAVVIAAPPTATDD30 pKa = 3.24SGDD33 pKa = 3.57AATHH37 pKa = 6.9DD38 pKa = 4.14SSQATDD44 pKa = 3.21SSQEE48 pKa = 3.85SGPAQEE54 pKa = 4.79SGVNVLVISSAVQDD68 pKa = 3.9PQSLQNAAKK77 pKa = 10.44DD78 pKa = 3.73GVKK81 pKa = 9.57TVFYY85 pKa = 10.22DD86 pKa = 3.68ANSTSLTEE94 pKa = 4.31LSQKK98 pKa = 9.46IANALNGQKK107 pKa = 10.33ADD109 pKa = 4.02SIAFAAEE116 pKa = 4.06GNDD119 pKa = 3.27NVLMLNTDD127 pKa = 2.87IMVSDD132 pKa = 5.21NIQSNIQNHH141 pKa = 7.06DD142 pKa = 2.79ISGFWKK148 pKa = 9.62TIGGLLNNGGRR159 pKa = 11.84IDD161 pKa = 5.72LLACDD166 pKa = 3.91VAGDD170 pKa = 3.79SGGQSLVAQIDD181 pKa = 3.63QLIDD185 pKa = 3.38RR186 pKa = 11.84PQDD189 pKa = 3.21GFFATVAASTDD200 pKa = 3.59LTGNAAGGDD209 pKa = 3.4WLLEE213 pKa = 4.07VGNIDD218 pKa = 4.1AKK220 pKa = 10.75SIYY223 pKa = 10.6FNDD226 pKa = 5.03SINNWNHH233 pKa = 4.62NMATAIEE240 pKa = 4.25YY241 pKa = 10.94GLIAALIGALLQPRR255 pKa = 11.84EE256 pKa = 4.17TLEE259 pKa = 4.11DD260 pKa = 3.34TSLSFSYY267 pKa = 10.65ASSGTVFANPDD278 pKa = 3.26FSFTLLTNSINGNTTVTEE296 pKa = 4.66DD297 pKa = 3.63GNVTYY302 pKa = 9.98TPNADD307 pKa = 3.52YY308 pKa = 11.2NNPGFTLGGTINDD321 pKa = 3.7GVSFDD326 pKa = 3.56SATIRR331 pKa = 11.84IVDD334 pKa = 4.24TNPDD338 pKa = 3.52DD339 pKa = 4.55PNSVFAINLPLYY351 pKa = 10.49LRR353 pKa = 11.84VVSVNDD359 pKa = 3.61APIAQNDD366 pKa = 4.05SFSIGQGNEE375 pKa = 3.49LTGNVALNDD384 pKa = 3.58SDD386 pKa = 4.16FQNGAPSEE394 pKa = 4.38NNTPLTVQLVSGPAHH409 pKa = 6.16AASFTLNADD418 pKa = 3.3GTFSYY423 pKa = 10.62TSIGSYY429 pKa = 10.24SGADD433 pKa = 2.95SFTYY437 pKa = 10.5SITDD441 pKa = 3.29SLGGVSEE448 pKa = 4.72ANVALSVISNAPPAVSDD465 pKa = 3.5NYY467 pKa = 10.29IINEE471 pKa = 3.95DD472 pKa = 3.39TTLNGNVTDD481 pKa = 4.34NDD483 pKa = 4.2TFNLGANQTIQLVSGPLVAANFALNSDD510 pKa = 4.01GSFSYY515 pKa = 10.59TPLANLVGVDD525 pKa = 3.28TFSYY529 pKa = 10.28VIKK532 pKa = 8.94TVSGAGITFSNTATVNIAVLPVNDD556 pKa = 4.17NPVAINDD563 pKa = 3.78NYY565 pKa = 10.94NVNEE569 pKa = 4.79DD570 pKa = 3.17GSLAANVLNNDD581 pKa = 2.98TDD583 pKa = 3.68IEE585 pKa = 4.37NNRR588 pKa = 11.84PFSAQLVAGPAHH600 pKa = 6.62AGSFTLNADD609 pKa = 3.43GTFSYY614 pKa = 10.69IPTADD619 pKa = 3.63YY620 pKa = 11.14NGSDD624 pKa = 3.11SFTYY628 pKa = 9.45VTVDD632 pKa = 3.05SLGAISNLATVSIAVASINDD652 pKa = 3.53APVAQDD658 pKa = 2.72VSYY661 pKa = 9.18NTNEE665 pKa = 4.02DD666 pKa = 3.75TPLTGILGVGSDD678 pKa = 4.2SQNGAPNEE686 pKa = 4.49SNTPLTAQLIGGPAHH701 pKa = 7.27ASAFTLNADD710 pKa = 3.67GSFSYY715 pKa = 10.85NPQADD720 pKa = 3.96FNGTDD725 pKa = 3.01SFTYY729 pKa = 10.27AIVDD733 pKa = 3.62SLGAISNTATVSITIDD749 pKa = 3.59SVNDD753 pKa = 3.36APIAQNDD760 pKa = 3.69AFQGNEE766 pKa = 3.96DD767 pKa = 3.47GSINGSVLLNDD778 pKa = 4.47SDD780 pKa = 4.27NQNGAPNEE788 pKa = 4.2NNSPLSAQLVTGPAHH803 pKa = 6.72AGSFTLNADD812 pKa = 3.38GTFTYY817 pKa = 10.23IPSADD822 pKa = 3.73YY823 pKa = 11.04NGSDD827 pKa = 2.98SFTYY831 pKa = 9.37VTLDD835 pKa = 3.29SLGGISNLATVSITINSVNDD855 pKa = 3.38APIAQNDD862 pKa = 4.16SYY864 pKa = 11.51QVNEE868 pKa = 4.2DD869 pKa = 3.18NGLNGNVALNDD880 pKa = 3.64SDD882 pKa = 4.23SQNGAPSEE890 pKa = 4.28NNGPLSAQLVTGPAHH905 pKa = 6.72AGSFTLNADD914 pKa = 3.43GTFSYY919 pKa = 10.62IPAANYY925 pKa = 10.17NGADD929 pKa = 3.15SFTYY933 pKa = 10.57VIVDD937 pKa = 3.55SLGATSNIATVAITINSVNDD957 pKa = 3.5APIVQNDD964 pKa = 3.61SFNIPGNLPYY974 pKa = 10.58SGNVLTNDD982 pKa = 3.68SDD984 pKa = 4.25NQGGAPNEE992 pKa = 4.0NNIPLSAQLVNGPAHH1007 pKa = 5.84ATSFILNADD1016 pKa = 3.64GTFSYY1021 pKa = 10.88QPANGFVGSDD1031 pKa = 2.88TFTYY1035 pKa = 10.4RR1036 pKa = 11.84VTDD1039 pKa = 3.64SLGGTSTIATVTITVAQAASANPILEE1065 pKa = 4.19LKK1067 pKa = 11.02GNINKK1072 pKa = 10.0NYY1074 pKa = 9.34IEE1076 pKa = 4.19NSGPVILSNNVKK1088 pKa = 8.65ITDD1091 pKa = 3.7SDD1093 pKa = 3.7SSNFDD1098 pKa = 3.08GGKK1101 pKa = 8.46LTVQIDD1107 pKa = 3.69QNGQSSDD1114 pKa = 3.06ILSIDD1119 pKa = 3.22NSTKK1123 pKa = 10.54VKK1125 pKa = 10.12TNANQDD1131 pKa = 2.6IYY1133 pKa = 11.73YY1134 pKa = 10.47NGLLVGHH1141 pKa = 5.5YY1142 pKa = 9.29TGGSGLASLEE1152 pKa = 3.74ITFNANATANVVSEE1166 pKa = 4.12VAEE1169 pKa = 4.42RR1170 pKa = 11.84IAYY1173 pKa = 8.97KK1174 pKa = 10.56NSSEE1178 pKa = 4.22NPSTADD1184 pKa = 3.33RR1185 pKa = 11.84SIRR1188 pKa = 11.84YY1189 pKa = 8.39QLSDD1193 pKa = 3.4GDD1195 pKa = 4.23SGISEE1200 pKa = 4.21AQYY1203 pKa = 10.56KK1204 pKa = 8.77IVRR1207 pKa = 11.84VFSVNDD1213 pKa = 3.54APDD1216 pKa = 3.06ISIADD1221 pKa = 3.44NTLTVNRR1228 pKa = 11.84GSITNIGALSGIQISDD1244 pKa = 3.19VDD1246 pKa = 3.69ASAGDD1251 pKa = 3.61RR1252 pKa = 11.84VRR1254 pKa = 11.84VVLKK1258 pKa = 10.84VLVGYY1263 pKa = 11.05LDD1265 pKa = 3.68FTDD1268 pKa = 5.09DD1269 pKa = 5.06LNDD1272 pKa = 3.36LTSIIFKK1279 pKa = 10.44RR1280 pKa = 11.84NPGDD1284 pKa = 3.49GLGSLDD1290 pKa = 3.84FTGTLADD1297 pKa = 3.4VNAALEE1303 pKa = 4.06RR1304 pKa = 11.84LTYY1307 pKa = 10.3RR1308 pKa = 11.84SRR1310 pKa = 11.84TNYY1313 pKa = 9.64IGLDD1317 pKa = 3.36ALTVGVSDD1325 pKa = 4.21MGSSGTFAIPRR1336 pKa = 11.84IDD1338 pKa = 4.99ADD1340 pKa = 3.53WLLLKK1345 pKa = 10.3IVV1347 pKa = 3.84
Molecular weight: 139.79 kDa
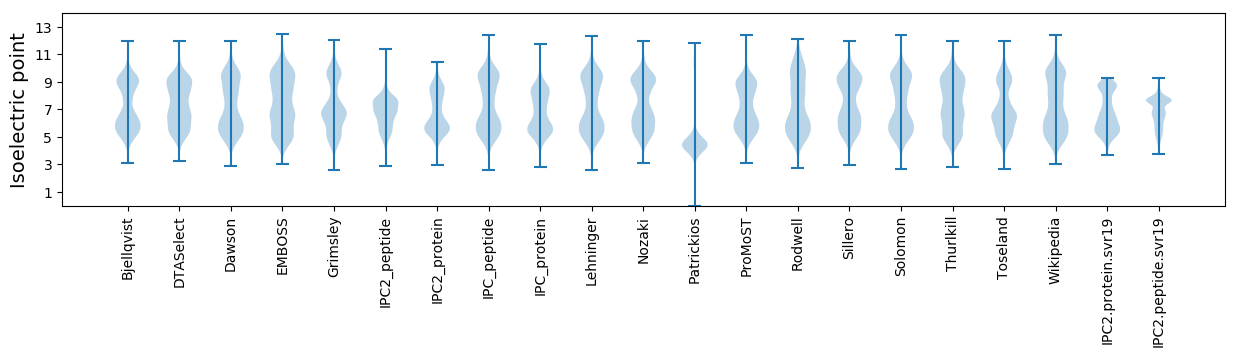
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A090D0X6|A0A090D0X6_9BACT Uncharacterized protein OS=Criblamydia sequanensis CRIB-18 OX=1437425 GN=CSEC_2205 PE=4 SV=1
MM1 pKa = 6.88LQEE4 pKa = 4.07YY5 pKa = 10.6VDD7 pKa = 3.23TGRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 7.4TAVASVRR19 pKa = 11.84LRR21 pKa = 11.84SGTGKK26 pKa = 10.33VDD28 pKa = 3.23INGKK32 pKa = 9.9AFEE35 pKa = 4.77EE36 pKa = 4.43YY37 pKa = 10.5FPLDD41 pKa = 3.62MQRR44 pKa = 11.84KK45 pKa = 8.27LALAPLGVVNVTGQFDD61 pKa = 3.87LLIRR65 pKa = 11.84VKK67 pKa = 10.86GGGVEE72 pKa = 4.39GQAEE76 pKa = 4.31AVRR79 pKa = 11.84LGLARR84 pKa = 11.84ALIQVNEE91 pKa = 4.06AFKK94 pKa = 11.25HH95 pKa = 4.43EE96 pKa = 4.37FKK98 pKa = 11.29VLGFLTRR105 pKa = 11.84DD106 pKa = 2.93SRR108 pKa = 11.84MKK110 pKa = 10.08EE111 pKa = 3.46RR112 pKa = 11.84KK113 pKa = 9.15KK114 pKa = 10.69YY115 pKa = 9.99GRR117 pKa = 11.84AGARR121 pKa = 11.84KK122 pKa = 9.15RR123 pKa = 11.84FQFSKK128 pKa = 10.78RR129 pKa = 3.42
MM1 pKa = 6.88LQEE4 pKa = 4.07YY5 pKa = 10.6VDD7 pKa = 3.23TGRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 7.4TAVASVRR19 pKa = 11.84LRR21 pKa = 11.84SGTGKK26 pKa = 10.33VDD28 pKa = 3.23INGKK32 pKa = 9.9AFEE35 pKa = 4.77EE36 pKa = 4.43YY37 pKa = 10.5FPLDD41 pKa = 3.62MQRR44 pKa = 11.84KK45 pKa = 8.27LALAPLGVVNVTGQFDD61 pKa = 3.87LLIRR65 pKa = 11.84VKK67 pKa = 10.86GGGVEE72 pKa = 4.39GQAEE76 pKa = 4.31AVRR79 pKa = 11.84LGLARR84 pKa = 11.84ALIQVNEE91 pKa = 4.06AFKK94 pKa = 11.25HH95 pKa = 4.43EE96 pKa = 4.37FKK98 pKa = 11.29VLGFLTRR105 pKa = 11.84DD106 pKa = 2.93SRR108 pKa = 11.84MKK110 pKa = 10.08EE111 pKa = 3.46RR112 pKa = 11.84KK113 pKa = 9.15KK114 pKa = 10.69YY115 pKa = 9.99GRR117 pKa = 11.84AGARR121 pKa = 11.84KK122 pKa = 9.15RR123 pKa = 11.84FQFSKK128 pKa = 10.78RR129 pKa = 3.42
Molecular weight: 14.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
885973 |
29 |
4164 |
366.4 |
41.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.536 ± 0.045 | 1.044 ± 0.022 |
4.893 ± 0.034 | 7.716 ± 0.05 |
5.626 ± 0.044 | 5.873 ± 0.055 |
1.999 ± 0.02 | 7.294 ± 0.039 |
8.269 ± 0.055 | 11.439 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.932 ± 0.025 | 4.555 ± 0.039 |
4.202 ± 0.04 | 3.288 ± 0.03 |
4.084 ± 0.032 | 7.23 ± 0.046 |
4.655 ± 0.032 | 5.159 ± 0.04 |
1.013 ± 0.017 | 3.194 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
