
Gillisia sp. Hel1_33_143
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Gillisia; unclassified Gillisia
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
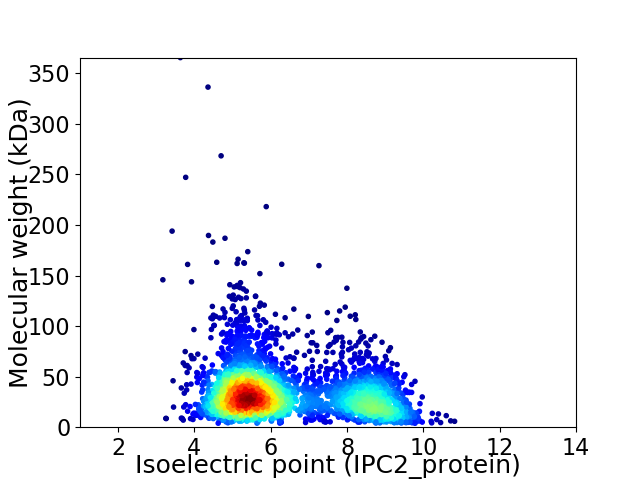
Virtual 2D-PAGE plot for 3141 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H1VGS7|A0A1H1VGS7_9FLAO Ammonium transporter OS=Gillisia sp. Hel1_33_143 OX=1336796 GN=SAMN04487764_3186 PE=3 SV=1
MM1 pKa = 7.64KK2 pKa = 10.4NFKK5 pKa = 10.43FLIAFAAMIILLFTYY20 pKa = 10.35CSKK23 pKa = 11.53DD24 pKa = 3.45DD25 pKa = 4.26VNDD28 pKa = 3.6QNSEE32 pKa = 4.33TNSEE36 pKa = 4.1LVSISFDD43 pKa = 3.5AVLKK47 pKa = 10.74DD48 pKa = 3.74FNTNRR53 pKa = 11.84SLTKK57 pKa = 10.19QNQLADD63 pKa = 3.46IPEE66 pKa = 4.54CLDD69 pKa = 4.09NIPSKK74 pKa = 11.14VRR76 pKa = 11.84VALKK80 pKa = 10.21DD81 pKa = 3.62SEE83 pKa = 4.56GAWVSMANGDD93 pKa = 3.42AGEE96 pKa = 4.52FIEE99 pKa = 5.61IPVIANDD106 pKa = 3.22EE107 pKa = 4.44GGWMTKK113 pKa = 9.66EE114 pKa = 4.11SADD117 pKa = 4.21LEE119 pKa = 4.63LPPGDD124 pKa = 3.5YY125 pKa = 9.9TLEE128 pKa = 4.02YY129 pKa = 10.26FAVLDD134 pKa = 3.68TDD136 pKa = 5.41NNVLWIAPRR145 pKa = 11.84EE146 pKa = 3.94NDD148 pKa = 4.22DD149 pKa = 3.6YY150 pKa = 12.04GPANFANFVSDD161 pKa = 4.69ALPITIDD168 pKa = 3.28LRR170 pKa = 11.84AGVKK174 pKa = 10.01KK175 pKa = 10.94YY176 pKa = 10.43IDD178 pKa = 3.99VEE180 pKa = 4.33VLCYY184 pKa = 9.61DD185 pKa = 3.22EE186 pKa = 6.3RR187 pKa = 11.84MADD190 pKa = 3.21EE191 pKa = 4.13YY192 pKa = 11.48GYY194 pKa = 11.4LFFDD198 pKa = 4.92FSQVDD203 pKa = 4.65AITLCLFGNYY213 pKa = 9.47CDD215 pKa = 3.57VTGRR219 pKa = 11.84HH220 pKa = 4.57YY221 pKa = 10.19PARR224 pKa = 11.84FSVEE228 pKa = 3.21AWIFSGDD235 pKa = 3.52LADD238 pKa = 4.74PKK240 pKa = 10.74LGEE243 pKa = 4.32PLTNGILTNEE253 pKa = 3.94TGIYY257 pKa = 10.17EE258 pKa = 4.16DD259 pKa = 4.47TEE261 pKa = 4.38DD262 pKa = 3.49AFARR266 pKa = 11.84PLCIALPNSSDD277 pKa = 3.04EE278 pKa = 3.81DD279 pKa = 3.84TYY281 pKa = 11.6YY282 pKa = 10.13IEE284 pKa = 4.68IKK286 pKa = 10.72LLGFEE291 pKa = 4.73GVYY294 pKa = 9.98TDD296 pKa = 3.93PEE298 pKa = 4.71VPVEE302 pKa = 3.99TLMITDD308 pKa = 4.13TEE310 pKa = 4.35VLDD313 pKa = 4.58LYY315 pKa = 11.07NGEE318 pKa = 4.49SNTYY322 pKa = 7.25YY323 pKa = 10.46HH324 pKa = 6.72FRR326 pKa = 11.84IGCEE330 pKa = 4.04GTEE333 pKa = 3.98EE334 pKa = 4.4CANDD338 pKa = 3.64MDD340 pKa = 5.6CDD342 pKa = 4.98GIADD346 pKa = 4.53DD347 pKa = 5.83VDD349 pKa = 3.81NCPMKK354 pKa = 11.05ANADD358 pKa = 3.65QTDD361 pKa = 3.46TDD363 pKa = 3.8EE364 pKa = 6.14DD365 pKa = 4.62GVGDD369 pKa = 3.98VCDD372 pKa = 3.54NCVNIANADD381 pKa = 3.79QVDD384 pKa = 3.66ADD386 pKa = 3.6QDD388 pKa = 4.44GIGDD392 pKa = 3.76ACEE395 pKa = 4.84DD396 pKa = 4.64LSTDD400 pKa = 3.87DD401 pKa = 6.01DD402 pKa = 5.26GDD404 pKa = 4.05GVPNDD409 pKa = 3.37IDD411 pKa = 3.91EE412 pKa = 4.99CPGTPSGTPVDD423 pKa = 3.47IKK425 pKa = 10.78GCEE428 pKa = 4.25SIQVPGRR435 pKa = 11.84DD436 pKa = 2.63VVVFNDD442 pKa = 4.05ANMFDD447 pKa = 4.49DD448 pKa = 5.67NAMEE452 pKa = 4.84DD453 pKa = 3.64PDD455 pKa = 3.9NIRR458 pKa = 11.84FVEE461 pKa = 4.07NLVTYY466 pKa = 7.59TTTGSRR472 pKa = 11.84NAGDD476 pKa = 3.81VVWIDD481 pKa = 3.38RR482 pKa = 11.84GRR484 pKa = 11.84SAACYY489 pKa = 10.69SNGEE493 pKa = 4.2CDD495 pKa = 3.31EE496 pKa = 5.26DD497 pKa = 3.69GWSKK501 pKa = 10.4MEE503 pKa = 3.83EE504 pKa = 4.39TITNTGLTVTSVFSNAGSLTNIPADD529 pKa = 3.44VKK531 pKa = 11.11VIFLVMPTLQYY542 pKa = 10.31TVAEE546 pKa = 4.19INAFKK551 pKa = 10.97AFAAEE556 pKa = 4.12GGRR559 pKa = 11.84IIFVGEE565 pKa = 3.63HH566 pKa = 5.09SSFYY570 pKa = 10.94FFIDD574 pKa = 3.31VQNQFLLNMGAVLTNTGGLVDD595 pKa = 4.5CGYY598 pKa = 9.08TVIPQSSNRR607 pKa = 11.84VHH609 pKa = 7.31PIMAGITDD617 pKa = 4.09LTIACASVIEE627 pKa = 4.4IGVDD631 pKa = 3.65DD632 pKa = 4.6FALFYY637 pKa = 9.97DD638 pKa = 4.01TTNTKK643 pKa = 10.0VLAGVAKK650 pKa = 10.13IDD652 pKa = 3.5TTPISEE658 pKa = 4.76LKK660 pKa = 10.51ASSIRR665 pKa = 11.84FDD667 pKa = 3.42NPVRR671 pKa = 11.84NRR673 pKa = 11.84NSKK676 pKa = 8.56STSSGIQPP684 pKa = 3.73
MM1 pKa = 7.64KK2 pKa = 10.4NFKK5 pKa = 10.43FLIAFAAMIILLFTYY20 pKa = 10.35CSKK23 pKa = 11.53DD24 pKa = 3.45DD25 pKa = 4.26VNDD28 pKa = 3.6QNSEE32 pKa = 4.33TNSEE36 pKa = 4.1LVSISFDD43 pKa = 3.5AVLKK47 pKa = 10.74DD48 pKa = 3.74FNTNRR53 pKa = 11.84SLTKK57 pKa = 10.19QNQLADD63 pKa = 3.46IPEE66 pKa = 4.54CLDD69 pKa = 4.09NIPSKK74 pKa = 11.14VRR76 pKa = 11.84VALKK80 pKa = 10.21DD81 pKa = 3.62SEE83 pKa = 4.56GAWVSMANGDD93 pKa = 3.42AGEE96 pKa = 4.52FIEE99 pKa = 5.61IPVIANDD106 pKa = 3.22EE107 pKa = 4.44GGWMTKK113 pKa = 9.66EE114 pKa = 4.11SADD117 pKa = 4.21LEE119 pKa = 4.63LPPGDD124 pKa = 3.5YY125 pKa = 9.9TLEE128 pKa = 4.02YY129 pKa = 10.26FAVLDD134 pKa = 3.68TDD136 pKa = 5.41NNVLWIAPRR145 pKa = 11.84EE146 pKa = 3.94NDD148 pKa = 4.22DD149 pKa = 3.6YY150 pKa = 12.04GPANFANFVSDD161 pKa = 4.69ALPITIDD168 pKa = 3.28LRR170 pKa = 11.84AGVKK174 pKa = 10.01KK175 pKa = 10.94YY176 pKa = 10.43IDD178 pKa = 3.99VEE180 pKa = 4.33VLCYY184 pKa = 9.61DD185 pKa = 3.22EE186 pKa = 6.3RR187 pKa = 11.84MADD190 pKa = 3.21EE191 pKa = 4.13YY192 pKa = 11.48GYY194 pKa = 11.4LFFDD198 pKa = 4.92FSQVDD203 pKa = 4.65AITLCLFGNYY213 pKa = 9.47CDD215 pKa = 3.57VTGRR219 pKa = 11.84HH220 pKa = 4.57YY221 pKa = 10.19PARR224 pKa = 11.84FSVEE228 pKa = 3.21AWIFSGDD235 pKa = 3.52LADD238 pKa = 4.74PKK240 pKa = 10.74LGEE243 pKa = 4.32PLTNGILTNEE253 pKa = 3.94TGIYY257 pKa = 10.17EE258 pKa = 4.16DD259 pKa = 4.47TEE261 pKa = 4.38DD262 pKa = 3.49AFARR266 pKa = 11.84PLCIALPNSSDD277 pKa = 3.04EE278 pKa = 3.81DD279 pKa = 3.84TYY281 pKa = 11.6YY282 pKa = 10.13IEE284 pKa = 4.68IKK286 pKa = 10.72LLGFEE291 pKa = 4.73GVYY294 pKa = 9.98TDD296 pKa = 3.93PEE298 pKa = 4.71VPVEE302 pKa = 3.99TLMITDD308 pKa = 4.13TEE310 pKa = 4.35VLDD313 pKa = 4.58LYY315 pKa = 11.07NGEE318 pKa = 4.49SNTYY322 pKa = 7.25YY323 pKa = 10.46HH324 pKa = 6.72FRR326 pKa = 11.84IGCEE330 pKa = 4.04GTEE333 pKa = 3.98EE334 pKa = 4.4CANDD338 pKa = 3.64MDD340 pKa = 5.6CDD342 pKa = 4.98GIADD346 pKa = 4.53DD347 pKa = 5.83VDD349 pKa = 3.81NCPMKK354 pKa = 11.05ANADD358 pKa = 3.65QTDD361 pKa = 3.46TDD363 pKa = 3.8EE364 pKa = 6.14DD365 pKa = 4.62GVGDD369 pKa = 3.98VCDD372 pKa = 3.54NCVNIANADD381 pKa = 3.79QVDD384 pKa = 3.66ADD386 pKa = 3.6QDD388 pKa = 4.44GIGDD392 pKa = 3.76ACEE395 pKa = 4.84DD396 pKa = 4.64LSTDD400 pKa = 3.87DD401 pKa = 6.01DD402 pKa = 5.26GDD404 pKa = 4.05GVPNDD409 pKa = 3.37IDD411 pKa = 3.91EE412 pKa = 4.99CPGTPSGTPVDD423 pKa = 3.47IKK425 pKa = 10.78GCEE428 pKa = 4.25SIQVPGRR435 pKa = 11.84DD436 pKa = 2.63VVVFNDD442 pKa = 4.05ANMFDD447 pKa = 4.49DD448 pKa = 5.67NAMEE452 pKa = 4.84DD453 pKa = 3.64PDD455 pKa = 3.9NIRR458 pKa = 11.84FVEE461 pKa = 4.07NLVTYY466 pKa = 7.59TTTGSRR472 pKa = 11.84NAGDD476 pKa = 3.81VVWIDD481 pKa = 3.38RR482 pKa = 11.84GRR484 pKa = 11.84SAACYY489 pKa = 10.69SNGEE493 pKa = 4.2CDD495 pKa = 3.31EE496 pKa = 5.26DD497 pKa = 3.69GWSKK501 pKa = 10.4MEE503 pKa = 3.83EE504 pKa = 4.39TITNTGLTVTSVFSNAGSLTNIPADD529 pKa = 3.44VKK531 pKa = 11.11VIFLVMPTLQYY542 pKa = 10.31TVAEE546 pKa = 4.19INAFKK551 pKa = 10.97AFAAEE556 pKa = 4.12GGRR559 pKa = 11.84IIFVGEE565 pKa = 3.63HH566 pKa = 5.09SSFYY570 pKa = 10.94FFIDD574 pKa = 3.31VQNQFLLNMGAVLTNTGGLVDD595 pKa = 4.5CGYY598 pKa = 9.08TVIPQSSNRR607 pKa = 11.84VHH609 pKa = 7.31PIMAGITDD617 pKa = 4.09LTIACASVIEE627 pKa = 4.4IGVDD631 pKa = 3.65DD632 pKa = 4.6FALFYY637 pKa = 9.97DD638 pKa = 4.01TTNTKK643 pKa = 10.0VLAGVAKK650 pKa = 10.13IDD652 pKa = 3.5TTPISEE658 pKa = 4.76LKK660 pKa = 10.51ASSIRR665 pKa = 11.84FDD667 pKa = 3.42NPVRR671 pKa = 11.84NRR673 pKa = 11.84NSKK676 pKa = 8.56STSSGIQPP684 pKa = 3.73
Molecular weight: 74.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H1SQJ5|A0A1H1SQJ5_9FLAO Uncharacterized protein OS=Gillisia sp. Hel1_33_143 OX=1336796 GN=SAMN04487764_2365 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1034711 |
34 |
3446 |
329.4 |
37.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.393 ± 0.036 | 0.687 ± 0.014 |
5.625 ± 0.04 | 6.935 ± 0.044 |
5.178 ± 0.038 | 6.243 ± 0.046 |
1.694 ± 0.023 | 8.155 ± 0.047 |
7.858 ± 0.056 | 9.565 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.287 ± 0.025 | 6.074 ± 0.048 |
3.304 ± 0.021 | 3.372 ± 0.024 |
3.503 ± 0.029 | 6.696 ± 0.035 |
5.419 ± 0.053 | 6.023 ± 0.038 |
0.99 ± 0.016 | 3.999 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
