
Pseudooceanicola lipolyticus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pseudooceanicola
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
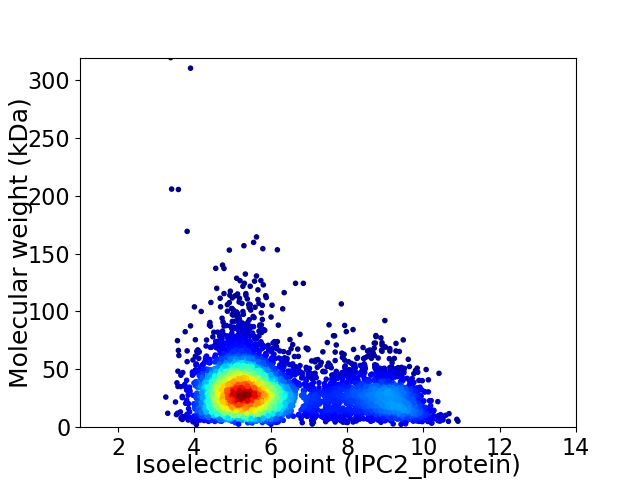
Virtual 2D-PAGE plot for 5156 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2M8IVR8|A0A2M8IVR8_9RHOB Corrinoid adenosyltransferase OS=Pseudooceanicola lipolyticus OX=2029104 GN=cobO PE=3 SV=1
MM1 pKa = 7.84KK2 pKa = 10.34YY3 pKa = 8.85LTFAAGVFGASAALAQTPEE22 pKa = 4.31LTVYY26 pKa = 10.09TYY28 pKa = 11.48DD29 pKa = 3.62SFVAEE34 pKa = 5.0WGPGPQIEE42 pKa = 4.53TAFEE46 pKa = 4.05AVCNCDD52 pKa = 3.27LTLVAAGDD60 pKa = 4.04GAALLARR67 pKa = 11.84LQLEE71 pKa = 4.35GDD73 pKa = 3.32RR74 pKa = 11.84TEE76 pKa = 4.96ADD78 pKa = 3.47VVLGLDD84 pKa = 3.72TNLVPAARR92 pKa = 11.84EE93 pKa = 3.87TGLFAPHH100 pKa = 6.6GADD103 pKa = 3.09ATLDD107 pKa = 4.42LPITWDD113 pKa = 3.99DD114 pKa = 3.91DD115 pKa = 3.86TFLPYY120 pKa = 10.65DD121 pKa = 3.12WGYY124 pKa = 10.36FAFIHH129 pKa = 5.55NTDD132 pKa = 4.14LANVPQDD139 pKa = 3.86FLEE142 pKa = 4.53LAASDD147 pKa = 4.19LSILIQDD154 pKa = 4.48PRR156 pKa = 11.84SSTPGLGLLMWVKK169 pKa = 10.34AAYY172 pKa = 9.95GDD174 pKa = 3.66EE175 pKa = 4.63APAVWAALADD185 pKa = 4.22NIVTVTKK192 pKa = 10.12GWSEE196 pKa = 4.19AYY198 pKa = 10.37GLFLDD203 pKa = 5.47GEE205 pKa = 4.66ADD207 pKa = 3.4MVLSYY212 pKa = 7.66TTSPAYY218 pKa = 10.17HH219 pKa = 7.21IIAEE223 pKa = 4.6DD224 pKa = 3.82DD225 pKa = 3.63TSKK228 pKa = 11.22DD229 pKa = 2.86AAAFDD234 pKa = 4.04EE235 pKa = 4.31GHH237 pKa = 6.05YY238 pKa = 10.6LQIEE242 pKa = 4.46LAAPLAGSDD251 pKa = 3.84QPEE254 pKa = 4.04LAAQFMQFITTDD266 pKa = 3.28AFQSVIPTTNWMYY279 pKa = 9.9PAVMPADD286 pKa = 4.04GLPAEE291 pKa = 4.94FDD293 pKa = 3.65TLVEE297 pKa = 4.17PRR299 pKa = 11.84NALLIPEE306 pKa = 4.58AEE308 pKa = 4.32VTEE311 pKa = 4.53LRR313 pKa = 11.84AQALAEE319 pKa = 3.99WLDD322 pKa = 3.67ALSQQ326 pKa = 3.47
MM1 pKa = 7.84KK2 pKa = 10.34YY3 pKa = 8.85LTFAAGVFGASAALAQTPEE22 pKa = 4.31LTVYY26 pKa = 10.09TYY28 pKa = 11.48DD29 pKa = 3.62SFVAEE34 pKa = 5.0WGPGPQIEE42 pKa = 4.53TAFEE46 pKa = 4.05AVCNCDD52 pKa = 3.27LTLVAAGDD60 pKa = 4.04GAALLARR67 pKa = 11.84LQLEE71 pKa = 4.35GDD73 pKa = 3.32RR74 pKa = 11.84TEE76 pKa = 4.96ADD78 pKa = 3.47VVLGLDD84 pKa = 3.72TNLVPAARR92 pKa = 11.84EE93 pKa = 3.87TGLFAPHH100 pKa = 6.6GADD103 pKa = 3.09ATLDD107 pKa = 4.42LPITWDD113 pKa = 3.99DD114 pKa = 3.91DD115 pKa = 3.86TFLPYY120 pKa = 10.65DD121 pKa = 3.12WGYY124 pKa = 10.36FAFIHH129 pKa = 5.55NTDD132 pKa = 4.14LANVPQDD139 pKa = 3.86FLEE142 pKa = 4.53LAASDD147 pKa = 4.19LSILIQDD154 pKa = 4.48PRR156 pKa = 11.84SSTPGLGLLMWVKK169 pKa = 10.34AAYY172 pKa = 9.95GDD174 pKa = 3.66EE175 pKa = 4.63APAVWAALADD185 pKa = 4.22NIVTVTKK192 pKa = 10.12GWSEE196 pKa = 4.19AYY198 pKa = 10.37GLFLDD203 pKa = 5.47GEE205 pKa = 4.66ADD207 pKa = 3.4MVLSYY212 pKa = 7.66TTSPAYY218 pKa = 10.17HH219 pKa = 7.21IIAEE223 pKa = 4.6DD224 pKa = 3.82DD225 pKa = 3.63TSKK228 pKa = 11.22DD229 pKa = 2.86AAAFDD234 pKa = 4.04EE235 pKa = 4.31GHH237 pKa = 6.05YY238 pKa = 10.6LQIEE242 pKa = 4.46LAAPLAGSDD251 pKa = 3.84QPEE254 pKa = 4.04LAAQFMQFITTDD266 pKa = 3.28AFQSVIPTTNWMYY279 pKa = 9.9PAVMPADD286 pKa = 4.04GLPAEE291 pKa = 4.94FDD293 pKa = 3.65TLVEE297 pKa = 4.17PRR299 pKa = 11.84NALLIPEE306 pKa = 4.58AEE308 pKa = 4.32VTEE311 pKa = 4.53LRR313 pKa = 11.84AQALAEE319 pKa = 3.99WLDD322 pKa = 3.67ALSQQ326 pKa = 3.47
Molecular weight: 35.07 kDa
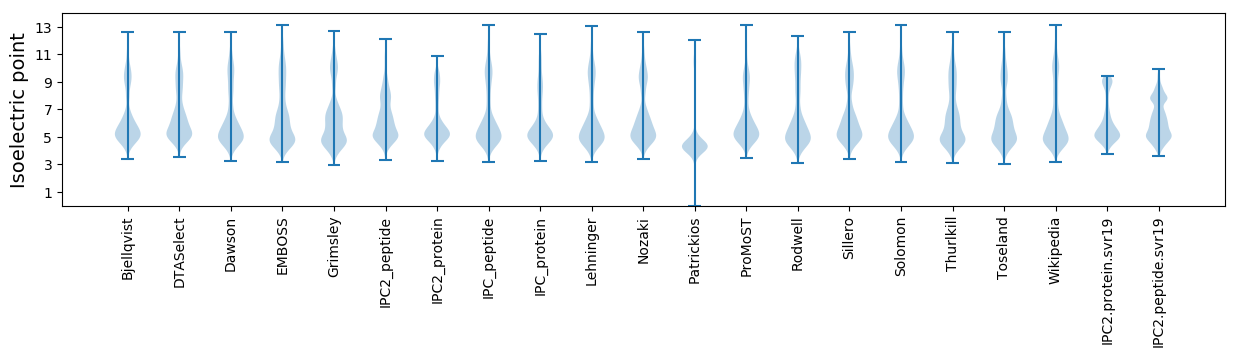
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2M8IYF9|A0A2M8IYF9_9RHOB sn-glycerol-3-phosphate ABC transporter ATP-binding protein UgpC OS=Pseudooceanicola lipolyticus OX=2029104 GN=CVM52_16705 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.43EE41 pKa = 3.72LSAA44 pKa = 5.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.43EE41 pKa = 3.72LSAA44 pKa = 5.03
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1548121 |
22 |
3102 |
300.3 |
32.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.644 ± 0.042 | 0.905 ± 0.011 |
5.991 ± 0.034 | 5.851 ± 0.035 |
3.736 ± 0.022 | 8.798 ± 0.037 |
2.016 ± 0.016 | 5.058 ± 0.022 |
2.83 ± 0.025 | 10.295 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.679 ± 0.018 | 2.482 ± 0.018 |
5.191 ± 0.025 | 3.294 ± 0.02 |
7.003 ± 0.035 | 5.088 ± 0.022 |
5.356 ± 0.026 | 7.193 ± 0.026 |
1.35 ± 0.013 | 2.24 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
