
Sulfuricella denitrificans (strain DSM 22764 / NBRC 105220 / skB26)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Sulfuricellaceae; Sulfuricella; Sulfuricella denitrificans
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
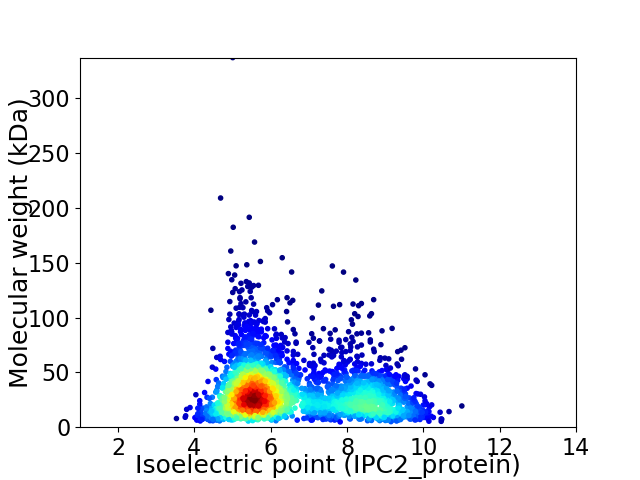
Virtual 2D-PAGE plot for 3071 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
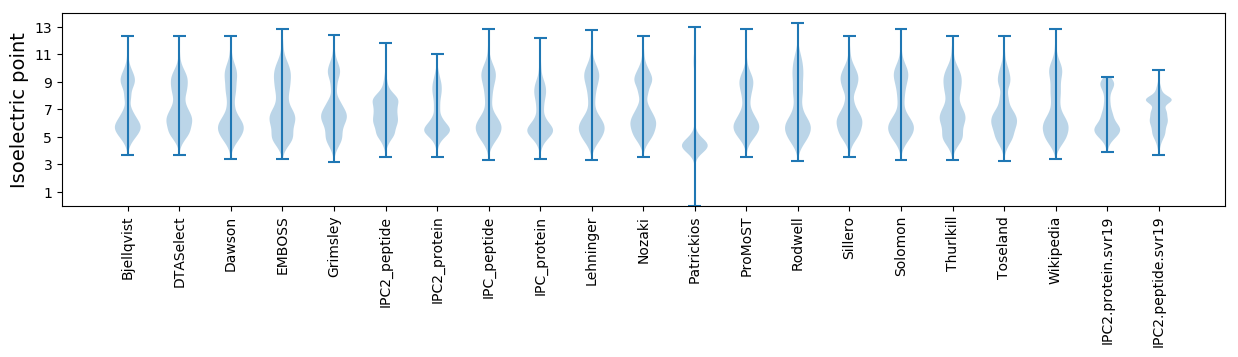
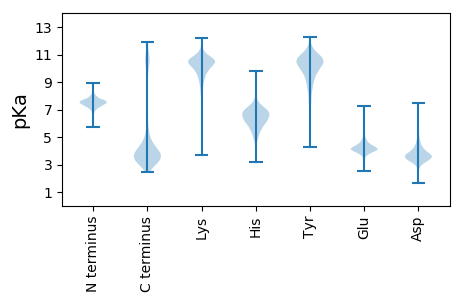
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S6AF13|S6AF13_SULDS UDP-N-acetylglucosamine 1-carboxyvinyltransferase OS=Sulfuricella denitrificans (strain DSM 22764 / NBRC 105220 / skB26) OX=1163617 GN=murA PE=3 SV=1
MM1 pKa = 7.62KK2 pKa = 10.51NNPLFKK8 pKa = 9.5TCLVAALVISGSVSTAQAATITFDD32 pKa = 4.53NLDD35 pKa = 3.9TSSGSYY41 pKa = 10.37AQLQNGYY48 pKa = 11.09AGFNWDD54 pKa = 4.1NFWAVDD60 pKa = 3.42GLNYY64 pKa = 10.1YY65 pKa = 10.32DD66 pKa = 5.05PNDD69 pKa = 3.72PTAIGYY75 pKa = 5.22TTGVVSPNNVAFNGFADD92 pKa = 4.07PASFSSTTIFSLEE105 pKa = 3.83SAYY108 pKa = 8.69VTKK111 pKa = 10.81ARR113 pKa = 11.84DD114 pKa = 3.14AGLTHH119 pKa = 7.04FDD121 pKa = 3.98GYY123 pKa = 11.52NGASLLYY130 pKa = 11.13SMDD133 pKa = 3.46VYY135 pKa = 11.05STTTAPTFVTFNWAGLDD152 pKa = 3.6KK153 pKa = 11.2VVMSDD158 pKa = 3.73GDD160 pKa = 4.16GTAHH164 pKa = 6.58TAIDD168 pKa = 5.27DD169 pKa = 3.68ITVNSVPVPAAAWLLGSGLLGLIGVARR196 pKa = 11.84RR197 pKa = 11.84KK198 pKa = 9.46VAA200 pKa = 3.33
MM1 pKa = 7.62KK2 pKa = 10.51NNPLFKK8 pKa = 9.5TCLVAALVISGSVSTAQAATITFDD32 pKa = 4.53NLDD35 pKa = 3.9TSSGSYY41 pKa = 10.37AQLQNGYY48 pKa = 11.09AGFNWDD54 pKa = 4.1NFWAVDD60 pKa = 3.42GLNYY64 pKa = 10.1YY65 pKa = 10.32DD66 pKa = 5.05PNDD69 pKa = 3.72PTAIGYY75 pKa = 5.22TTGVVSPNNVAFNGFADD92 pKa = 4.07PASFSSTTIFSLEE105 pKa = 3.83SAYY108 pKa = 8.69VTKK111 pKa = 10.81ARR113 pKa = 11.84DD114 pKa = 3.14AGLTHH119 pKa = 7.04FDD121 pKa = 3.98GYY123 pKa = 11.52NGASLLYY130 pKa = 11.13SMDD133 pKa = 3.46VYY135 pKa = 11.05STTTAPTFVTFNWAGLDD152 pKa = 3.6KK153 pKa = 11.2VVMSDD158 pKa = 3.73GDD160 pKa = 4.16GTAHH164 pKa = 6.58TAIDD168 pKa = 5.27DD169 pKa = 3.68ITVNSVPVPAAAWLLGSGLLGLIGVARR196 pKa = 11.84RR197 pKa = 11.84KK198 pKa = 9.46VAA200 pKa = 3.33
Molecular weight: 21.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S6AMC0|S6AMC0_SULDS Type IV pilus assembly protein PilW OS=Sulfuricella denitrificans (strain DSM 22764 / NBRC 105220 / skB26) OX=1163617 GN=SCD_n02125 PE=4 SV=1
MM1 pKa = 7.25VSSLNRR7 pKa = 11.84GFVIFAVLCAASWTTAVFATSCRR30 pKa = 11.84EE31 pKa = 3.62AAHH34 pKa = 6.84ALNQRR39 pKa = 11.84LQPKK43 pKa = 9.49IDD45 pKa = 3.63EE46 pKa = 4.73AEE48 pKa = 4.13LANMLDD54 pKa = 3.57VLNEE58 pKa = 3.94SRR60 pKa = 11.84NTQLPGKK67 pKa = 9.79FVTKK71 pKa = 10.44RR72 pKa = 11.84QAQQSGWRR80 pKa = 11.84PGRR83 pKa = 11.84DD84 pKa = 3.19LWNVPALRR92 pKa = 11.84FKK94 pKa = 11.13SIGGDD99 pKa = 2.82HH100 pKa = 6.88FGNRR104 pKa = 11.84EE105 pKa = 3.44GRR107 pKa = 11.84LPRR110 pKa = 11.84GDD112 pKa = 3.12WRR114 pKa = 11.84EE115 pKa = 3.08ADD117 pKa = 3.22LNYY120 pKa = 10.61RR121 pKa = 11.84GGHH124 pKa = 5.94RR125 pKa = 11.84GAKK128 pKa = 9.2RR129 pKa = 11.84LLFSRR134 pKa = 11.84EE135 pKa = 3.28GLRR138 pKa = 11.84RR139 pKa = 11.84VTVDD143 pKa = 3.17HH144 pKa = 6.23YY145 pKa = 9.96QTFVEE150 pKa = 4.93VPPCRR155 pKa = 3.66
MM1 pKa = 7.25VSSLNRR7 pKa = 11.84GFVIFAVLCAASWTTAVFATSCRR30 pKa = 11.84EE31 pKa = 3.62AAHH34 pKa = 6.84ALNQRR39 pKa = 11.84LQPKK43 pKa = 9.49IDD45 pKa = 3.63EE46 pKa = 4.73AEE48 pKa = 4.13LANMLDD54 pKa = 3.57VLNEE58 pKa = 3.94SRR60 pKa = 11.84NTQLPGKK67 pKa = 9.79FVTKK71 pKa = 10.44RR72 pKa = 11.84QAQQSGWRR80 pKa = 11.84PGRR83 pKa = 11.84DD84 pKa = 3.19LWNVPALRR92 pKa = 11.84FKK94 pKa = 11.13SIGGDD99 pKa = 2.82HH100 pKa = 6.88FGNRR104 pKa = 11.84EE105 pKa = 3.44GRR107 pKa = 11.84LPRR110 pKa = 11.84GDD112 pKa = 3.12WRR114 pKa = 11.84EE115 pKa = 3.08ADD117 pKa = 3.22LNYY120 pKa = 10.61RR121 pKa = 11.84GGHH124 pKa = 5.94RR125 pKa = 11.84GAKK128 pKa = 9.2RR129 pKa = 11.84LLFSRR134 pKa = 11.84EE135 pKa = 3.28GLRR138 pKa = 11.84RR139 pKa = 11.84VTVDD143 pKa = 3.17HH144 pKa = 6.23YY145 pKa = 9.96QTFVEE150 pKa = 4.93VPPCRR155 pKa = 3.66
Molecular weight: 17.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
966249 |
46 |
3317 |
314.6 |
34.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.374 ± 0.053 | 0.991 ± 0.021 |
5.225 ± 0.033 | 6.146 ± 0.052 |
3.81 ± 0.03 | 7.764 ± 0.047 |
2.386 ± 0.023 | 5.549 ± 0.03 |
4.517 ± 0.041 | 10.905 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.674 ± 0.024 | 3.308 ± 0.028 |
4.649 ± 0.03 | 3.901 ± 0.03 |
5.973 ± 0.043 | 5.696 ± 0.039 |
5.053 ± 0.036 | 7.176 ± 0.044 |
1.278 ± 0.017 | 2.625 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
