
Tardiphaga robiniae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Bradyrhizobiaceae; Tardiphaga
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
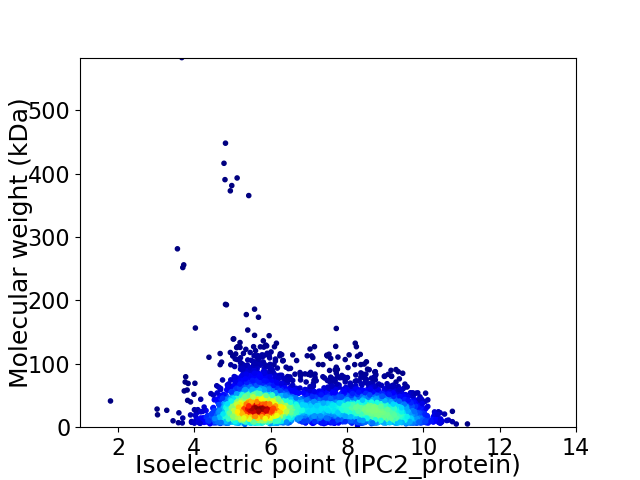
Virtual 2D-PAGE plot for 5721 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A164ART6|A0A164ART6_9BRAD Corrinoid adenosyltransferase OS=Tardiphaga robiniae OX=943830 GN=A4A58_01690 PE=3 SV=1
MM1 pKa = 7.35SLSGALSSAISALNAQSSALAMISDD26 pKa = 5.05NIANSSTVGYY36 pKa = 7.67KK37 pKa = 7.36TTSASFEE44 pKa = 4.42SLVTSSSSASSYY56 pKa = 11.11SSGGVSTLSKK66 pKa = 11.28SNITTQGLLTATSTSTNIAIQGNGFFPVTTSLTGGSTLYY105 pKa = 9.85TRR107 pKa = 11.84NGAFTTDD114 pKa = 2.73SDD116 pKa = 4.61GYY118 pKa = 10.73LVNNGAYY125 pKa = 10.56LLGWRR130 pKa = 11.84TDD132 pKa = 2.87ASGNVIGSASSGNLTAIDD150 pKa = 3.53TSVAQTSGSATTTTTFSANLPADD173 pKa = 3.68AAVGDD178 pKa = 4.15TFTTSMSAYY187 pKa = 9.86DD188 pKa = 3.61SLGTSSSIQVTWSKK202 pKa = 9.2TADD205 pKa = 3.79NTWSASFGQPTLSSDD220 pKa = 3.73STVTTAATPTSTVAVTFNTDD240 pKa = 2.38GTLASTTPDD249 pKa = 3.67PPVLALTGSTTGAADD264 pKa = 3.37NTITLNLGTVGGTDD278 pKa = 3.53GLTQYY283 pKa = 11.65ASGLSTPAVNITRR296 pKa = 11.84IEE298 pKa = 4.41SNGLAFGKK306 pKa = 10.25LSNISVGDD314 pKa = 3.65NGVVNATYY322 pKa = 11.24SNGQTIAIYY331 pKa = 9.49KK332 pKa = 10.05IPVATFAAADD342 pKa = 3.57QLEE345 pKa = 4.34AQSNGLYY352 pKa = 9.21STTAEE357 pKa = 4.3SGTAALQEE365 pKa = 4.55SGTNGAGTIYY375 pKa = 10.5GSEE378 pKa = 4.49LEE380 pKa = 4.75SSTTDD385 pKa = 3.06TNEE388 pKa = 3.87QFSSMISAQQAYY400 pKa = 9.56SAASQVITAVNKK412 pKa = 9.85MFDD415 pKa = 3.61TLISSMRR422 pKa = 3.49
MM1 pKa = 7.35SLSGALSSAISALNAQSSALAMISDD26 pKa = 5.05NIANSSTVGYY36 pKa = 7.67KK37 pKa = 7.36TTSASFEE44 pKa = 4.42SLVTSSSSASSYY56 pKa = 11.11SSGGVSTLSKK66 pKa = 11.28SNITTQGLLTATSTSTNIAIQGNGFFPVTTSLTGGSTLYY105 pKa = 9.85TRR107 pKa = 11.84NGAFTTDD114 pKa = 2.73SDD116 pKa = 4.61GYY118 pKa = 10.73LVNNGAYY125 pKa = 10.56LLGWRR130 pKa = 11.84TDD132 pKa = 2.87ASGNVIGSASSGNLTAIDD150 pKa = 3.53TSVAQTSGSATTTTTFSANLPADD173 pKa = 3.68AAVGDD178 pKa = 4.15TFTTSMSAYY187 pKa = 9.86DD188 pKa = 3.61SLGTSSSIQVTWSKK202 pKa = 9.2TADD205 pKa = 3.79NTWSASFGQPTLSSDD220 pKa = 3.73STVTTAATPTSTVAVTFNTDD240 pKa = 2.38GTLASTTPDD249 pKa = 3.67PPVLALTGSTTGAADD264 pKa = 3.37NTITLNLGTVGGTDD278 pKa = 3.53GLTQYY283 pKa = 11.65ASGLSTPAVNITRR296 pKa = 11.84IEE298 pKa = 4.41SNGLAFGKK306 pKa = 10.25LSNISVGDD314 pKa = 3.65NGVVNATYY322 pKa = 11.24SNGQTIAIYY331 pKa = 9.49KK332 pKa = 10.05IPVATFAAADD342 pKa = 3.57QLEE345 pKa = 4.34AQSNGLYY352 pKa = 9.21STTAEE357 pKa = 4.3SGTAALQEE365 pKa = 4.55SGTNGAGTIYY375 pKa = 10.5GSEE378 pKa = 4.49LEE380 pKa = 4.75SSTTDD385 pKa = 3.06TNEE388 pKa = 3.87QFSSMISAQQAYY400 pKa = 9.56SAASQVITAVNKK412 pKa = 9.85MFDD415 pKa = 3.61TLISSMRR422 pKa = 3.49
Molecular weight: 42.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A163ZA08|A0A163ZA08_9BRAD Class II aldolase OS=Tardiphaga robiniae OX=943830 GN=A4A58_07385 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATVGGRR28 pKa = 11.84KK29 pKa = 9.08VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATVGGRR28 pKa = 11.84KK29 pKa = 9.08VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1807327 |
26 |
5746 |
315.9 |
34.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.217 ± 0.038 | 0.78 ± 0.011 |
5.666 ± 0.032 | 4.989 ± 0.035 |
3.8 ± 0.021 | 8.467 ± 0.049 |
1.924 ± 0.019 | 5.658 ± 0.023 |
3.872 ± 0.03 | 9.766 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.605 ± 0.018 | 2.933 ± 0.026 |
5.073 ± 0.026 | 3.185 ± 0.018 |
6.401 ± 0.037 | 5.79 ± 0.028 |
5.705 ± 0.037 | 7.611 ± 0.027 |
1.269 ± 0.012 | 2.289 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
