
Methanobrevibacter woesei
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Methanomada group; Methanobacteria; Methanobacteriales; Methanobacteriaceae; Methanobrevibacter
Average proteome isoelectric point is 5.63
Get precalculated fractions of proteins
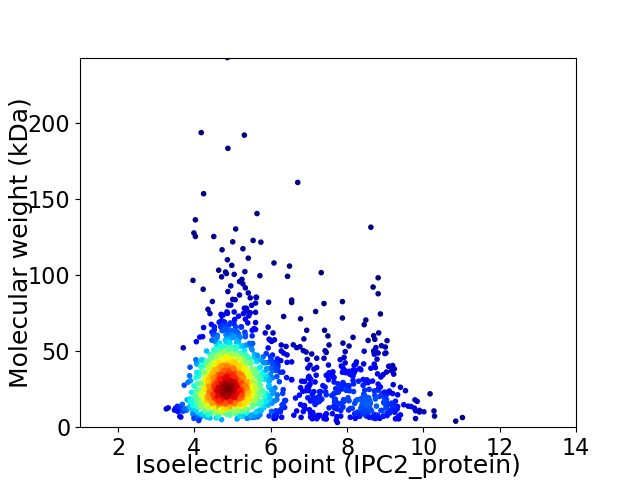
Virtual 2D-PAGE plot for 1581 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U1S9R8|A0A2U1S9R8_9EURY Uncharacterized protein OS=Methanobrevibacter woesei OX=190976 GN=MBBWO_02570 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84KK3 pKa = 9.41AVIGIIVVVILIAGGVGLSANSINNGNNNSTVITYY38 pKa = 11.0GEE40 pKa = 4.05TTFANDD46 pKa = 3.17QYY48 pKa = 11.64KK49 pKa = 10.73SLVDD53 pKa = 3.61SYY55 pKa = 11.83FMNNNYY61 pKa = 10.26GNLEE65 pKa = 3.98SMNYY69 pKa = 10.1KK70 pKa = 10.2IITADD75 pKa = 3.3EE76 pKa = 4.29VNAISQGISQEE87 pKa = 4.56TYY89 pKa = 9.91SSGQIFSSSLVDD101 pKa = 4.18LGSSEE106 pKa = 4.76DD107 pKa = 3.57LSIDD111 pKa = 3.32VDD113 pKa = 3.62HH114 pKa = 7.27SKK116 pKa = 8.56ITLVTDD122 pKa = 3.9DD123 pKa = 4.36MYY125 pKa = 11.69KK126 pKa = 10.34SALEE130 pKa = 3.97STGITKK136 pKa = 9.98GHH138 pKa = 6.51VIVTSPVSATGEE150 pKa = 4.08SALAGIMSAYY160 pKa = 9.85EE161 pKa = 3.77YY162 pKa = 9.68ATNTEE167 pKa = 3.69IPEE170 pKa = 4.27EE171 pKa = 4.1VKK173 pKa = 10.51EE174 pKa = 4.02AANEE178 pKa = 4.15EE179 pKa = 4.7IYY181 pKa = 8.06TTSEE185 pKa = 3.46IVNNSNVSADD195 pKa = 3.83DD196 pKa = 3.55LTDD199 pKa = 3.33VVDD202 pKa = 4.03EE203 pKa = 4.41VKK205 pKa = 11.09EE206 pKa = 3.9EE207 pKa = 4.13VEE209 pKa = 3.97AQNITNHH216 pKa = 4.74EE217 pKa = 4.39TIVNIINNVTVNYY230 pKa = 10.37NIVLSPEE237 pKa = 4.59DD238 pKa = 3.64IEE240 pKa = 5.03NLAEE244 pKa = 5.15SIRR247 pKa = 11.84QTQLVQDD254 pKa = 3.8QASDD258 pKa = 3.7YY259 pKa = 10.57QSQINSFIQNNTGGFSLEE277 pKa = 4.31NIFNFF282 pKa = 4.37
MM1 pKa = 7.62RR2 pKa = 11.84KK3 pKa = 9.41AVIGIIVVVILIAGGVGLSANSINNGNNNSTVITYY38 pKa = 11.0GEE40 pKa = 4.05TTFANDD46 pKa = 3.17QYY48 pKa = 11.64KK49 pKa = 10.73SLVDD53 pKa = 3.61SYY55 pKa = 11.83FMNNNYY61 pKa = 10.26GNLEE65 pKa = 3.98SMNYY69 pKa = 10.1KK70 pKa = 10.2IITADD75 pKa = 3.3EE76 pKa = 4.29VNAISQGISQEE87 pKa = 4.56TYY89 pKa = 9.91SSGQIFSSSLVDD101 pKa = 4.18LGSSEE106 pKa = 4.76DD107 pKa = 3.57LSIDD111 pKa = 3.32VDD113 pKa = 3.62HH114 pKa = 7.27SKK116 pKa = 8.56ITLVTDD122 pKa = 3.9DD123 pKa = 4.36MYY125 pKa = 11.69KK126 pKa = 10.34SALEE130 pKa = 3.97STGITKK136 pKa = 9.98GHH138 pKa = 6.51VIVTSPVSATGEE150 pKa = 4.08SALAGIMSAYY160 pKa = 9.85EE161 pKa = 3.77YY162 pKa = 9.68ATNTEE167 pKa = 3.69IPEE170 pKa = 4.27EE171 pKa = 4.1VKK173 pKa = 10.51EE174 pKa = 4.02AANEE178 pKa = 4.15EE179 pKa = 4.7IYY181 pKa = 8.06TTSEE185 pKa = 3.46IVNNSNVSADD195 pKa = 3.83DD196 pKa = 3.55LTDD199 pKa = 3.33VVDD202 pKa = 4.03EE203 pKa = 4.41VKK205 pKa = 11.09EE206 pKa = 3.9EE207 pKa = 4.13VEE209 pKa = 3.97AQNITNHH216 pKa = 4.74EE217 pKa = 4.39TIVNIINNVTVNYY230 pKa = 10.37NIVLSPEE237 pKa = 4.59DD238 pKa = 3.64IEE240 pKa = 5.03NLAEE244 pKa = 5.15SIRR247 pKa = 11.84QTQLVQDD254 pKa = 3.8QASDD258 pKa = 3.7YY259 pKa = 10.57QSQINSFIQNNTGGFSLEE277 pKa = 4.31NIFNFF282 pKa = 4.37
Molecular weight: 30.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U1S6E9|A0A2U1S6E9_9EURY Copper-exporting P-type ATPase A OS=Methanobrevibacter woesei OX=190976 GN=copA_1 PE=3 SV=1
MM1 pKa = 7.51SRR3 pKa = 11.84NKK5 pKa = 9.92PLAKK9 pKa = 9.48KK10 pKa = 10.5LRR12 pKa = 11.84MAKK15 pKa = 10.13ANKK18 pKa = 7.84QNRR21 pKa = 11.84RR22 pKa = 11.84IPIWAYY28 pKa = 10.35AKK30 pKa = 9.41TNRR33 pKa = 11.84KK34 pKa = 8.76LRR36 pKa = 11.84YY37 pKa = 8.95RR38 pKa = 11.84PKK40 pKa = 8.62PRR42 pKa = 11.84HH43 pKa = 4.36WRR45 pKa = 11.84RR46 pKa = 11.84NSLKK50 pKa = 10.73LL51 pKa = 3.43
MM1 pKa = 7.51SRR3 pKa = 11.84NKK5 pKa = 9.92PLAKK9 pKa = 9.48KK10 pKa = 10.5LRR12 pKa = 11.84MAKK15 pKa = 10.13ANKK18 pKa = 7.84QNRR21 pKa = 11.84RR22 pKa = 11.84IPIWAYY28 pKa = 10.35AKK30 pKa = 9.41TNRR33 pKa = 11.84KK34 pKa = 8.76LRR36 pKa = 11.84YY37 pKa = 8.95RR38 pKa = 11.84PKK40 pKa = 8.62PRR42 pKa = 11.84HH43 pKa = 4.36WRR45 pKa = 11.84RR46 pKa = 11.84NSLKK50 pKa = 10.73LL51 pKa = 3.43
Molecular weight: 6.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
459352 |
29 |
2108 |
290.5 |
32.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.005 ± 0.072 | 1.294 ± 0.033 |
6.582 ± 0.05 | 7.98 ± 0.072 |
4.321 ± 0.055 | 6.417 ± 0.07 |
1.603 ± 0.025 | 9.529 ± 0.068 |
7.985 ± 0.074 | 8.846 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.384 ± 0.03 | 6.019 ± 0.083 |
3.374 ± 0.035 | 2.175 ± 0.026 |
3.239 ± 0.044 | 6.32 ± 0.052 |
4.868 ± 0.044 | 6.642 ± 0.063 |
0.629 ± 0.016 | 3.791 ± 0.05 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
