
Achlya hypogyna
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Saprolegniales; Saprolegniaceae; Achlya
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
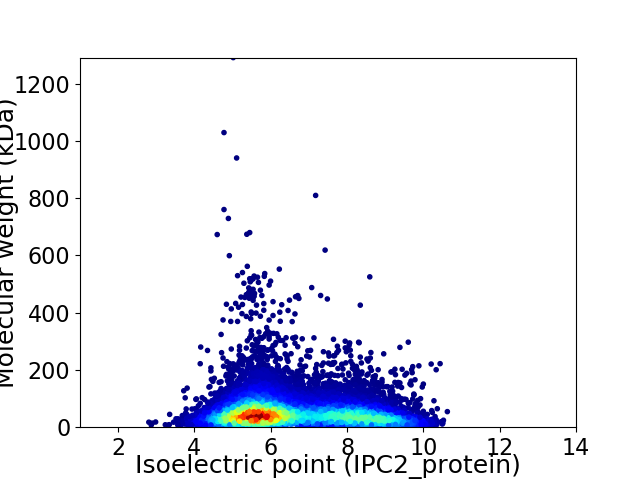
Virtual 2D-PAGE plot for 14400 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V9Y4U3|A0A1V9Y4U3_9STRA Uncharacterized protein OS=Achlya hypogyna OX=1202772 GN=ACHHYP_17267 PE=4 SV=1
MM1 pKa = 7.17VFLPFVLLALSGVGAAGIALRR22 pKa = 11.84SCCYY26 pKa = 10.22KK27 pKa = 10.75PEE29 pKa = 4.43DD30 pKa = 3.48EE31 pKa = 4.51WRR33 pKa = 11.84DD34 pKa = 3.52PRR36 pKa = 11.84QEE38 pKa = 4.26PLMEE42 pKa = 4.84APNVPATTKK51 pKa = 10.75ASAPSVTPTLAPPPVVKK68 pKa = 10.62SSTPPPTAVVKK79 pKa = 10.37PLDD82 pKa = 4.1GVGGADD88 pKa = 3.53PMVEE92 pKa = 3.87PVACRR97 pKa = 11.84GCGATIDD104 pKa = 4.04PSLAALANSLCIVCSYY120 pKa = 10.88QSIRR124 pKa = 11.84LPSLEE129 pKa = 4.5IEE131 pKa = 4.42QPDD134 pKa = 3.74DD135 pKa = 3.44TFPEE139 pKa = 4.55YY140 pKa = 10.18IANDD144 pKa = 4.03DD145 pKa = 4.83ADD147 pKa = 5.43DD148 pKa = 4.75DD149 pKa = 4.25MDD151 pKa = 4.41GAGYY155 pKa = 10.41DD156 pKa = 4.12DD157 pKa = 5.53DD158 pKa = 3.44MSYY161 pKa = 11.2YY162 pKa = 10.73YY163 pKa = 10.28KK164 pKa = 10.91DD165 pKa = 3.21KK166 pKa = 11.34ARR168 pKa = 11.84TTDD171 pKa = 3.04EE172 pKa = 4.01GTTAEE177 pKa = 4.37MEE179 pKa = 4.47SPQDD183 pKa = 3.92LDD185 pKa = 5.82DD186 pKa = 5.16GALDD190 pKa = 6.08DD191 pKa = 6.11DD192 pKa = 5.58DD193 pKa = 6.19EE194 pKa = 7.59DD195 pKa = 4.57DD196 pKa = 5.21ASSLSQEE203 pKa = 4.1MEE205 pKa = 3.98IPCADD210 pKa = 3.92GEE212 pKa = 4.28PAADD216 pKa = 3.79EE217 pKa = 5.0DD218 pKa = 4.47IEE220 pKa = 4.26IAEE223 pKa = 4.21EE224 pKa = 3.97ALALVQDD231 pKa = 3.99MWDD234 pKa = 3.05IAYY237 pKa = 9.06QAHH240 pKa = 6.91LGAGEE245 pKa = 4.66GPDD248 pKa = 2.94GDD250 pKa = 4.49AFVEE254 pKa = 4.3LALEE258 pKa = 4.51LDD260 pKa = 3.42ATAAAIVKK268 pKa = 9.3EE269 pKa = 3.96PHH271 pKa = 6.21LLSEE275 pKa = 4.53SFHH278 pKa = 6.65FLSQALASLLEE289 pKa = 4.3LVPEE293 pKa = 4.54AWAPHH298 pKa = 5.25VEE300 pKa = 3.87ATEE303 pKa = 4.26LKK305 pKa = 10.69FEE307 pKa = 4.38ALQFRR312 pKa = 11.84YY313 pKa = 9.46HH314 pKa = 6.67SKK316 pKa = 9.31LTVEE320 pKa = 4.89CGLDD324 pKa = 3.45LAAQLYY330 pKa = 9.81EE331 pKa = 4.16LVEE334 pKa = 4.36CASDD338 pKa = 3.73FGVDD342 pKa = 3.37PTVAASVLQGLEE354 pKa = 3.96EE355 pKa = 3.77LAGAVEE361 pKa = 4.26EE362 pKa = 4.89TPCEE366 pKa = 3.84LVSWLNYY373 pKa = 10.19LGATIKK379 pKa = 10.6LLKK382 pKa = 10.19SYY384 pKa = 9.85EE385 pKa = 4.06RR386 pKa = 11.84EE387 pKa = 4.07FEE389 pKa = 5.79DD390 pKa = 3.6VAKK393 pKa = 10.2WDD395 pKa = 3.99DD396 pKa = 3.62VVDD399 pKa = 4.26CEE401 pKa = 4.7RR402 pKa = 11.84HH403 pKa = 5.72LEE405 pKa = 3.89PLQLHH410 pKa = 6.15CWEE413 pKa = 5.16IYY415 pKa = 10.2SPCC418 pKa = 4.06
MM1 pKa = 7.17VFLPFVLLALSGVGAAGIALRR22 pKa = 11.84SCCYY26 pKa = 10.22KK27 pKa = 10.75PEE29 pKa = 4.43DD30 pKa = 3.48EE31 pKa = 4.51WRR33 pKa = 11.84DD34 pKa = 3.52PRR36 pKa = 11.84QEE38 pKa = 4.26PLMEE42 pKa = 4.84APNVPATTKK51 pKa = 10.75ASAPSVTPTLAPPPVVKK68 pKa = 10.62SSTPPPTAVVKK79 pKa = 10.37PLDD82 pKa = 4.1GVGGADD88 pKa = 3.53PMVEE92 pKa = 3.87PVACRR97 pKa = 11.84GCGATIDD104 pKa = 4.04PSLAALANSLCIVCSYY120 pKa = 10.88QSIRR124 pKa = 11.84LPSLEE129 pKa = 4.5IEE131 pKa = 4.42QPDD134 pKa = 3.74DD135 pKa = 3.44TFPEE139 pKa = 4.55YY140 pKa = 10.18IANDD144 pKa = 4.03DD145 pKa = 4.83ADD147 pKa = 5.43DD148 pKa = 4.75DD149 pKa = 4.25MDD151 pKa = 4.41GAGYY155 pKa = 10.41DD156 pKa = 4.12DD157 pKa = 5.53DD158 pKa = 3.44MSYY161 pKa = 11.2YY162 pKa = 10.73YY163 pKa = 10.28KK164 pKa = 10.91DD165 pKa = 3.21KK166 pKa = 11.34ARR168 pKa = 11.84TTDD171 pKa = 3.04EE172 pKa = 4.01GTTAEE177 pKa = 4.37MEE179 pKa = 4.47SPQDD183 pKa = 3.92LDD185 pKa = 5.82DD186 pKa = 5.16GALDD190 pKa = 6.08DD191 pKa = 6.11DD192 pKa = 5.58DD193 pKa = 6.19EE194 pKa = 7.59DD195 pKa = 4.57DD196 pKa = 5.21ASSLSQEE203 pKa = 4.1MEE205 pKa = 3.98IPCADD210 pKa = 3.92GEE212 pKa = 4.28PAADD216 pKa = 3.79EE217 pKa = 5.0DD218 pKa = 4.47IEE220 pKa = 4.26IAEE223 pKa = 4.21EE224 pKa = 3.97ALALVQDD231 pKa = 3.99MWDD234 pKa = 3.05IAYY237 pKa = 9.06QAHH240 pKa = 6.91LGAGEE245 pKa = 4.66GPDD248 pKa = 2.94GDD250 pKa = 4.49AFVEE254 pKa = 4.3LALEE258 pKa = 4.51LDD260 pKa = 3.42ATAAAIVKK268 pKa = 9.3EE269 pKa = 3.96PHH271 pKa = 6.21LLSEE275 pKa = 4.53SFHH278 pKa = 6.65FLSQALASLLEE289 pKa = 4.3LVPEE293 pKa = 4.54AWAPHH298 pKa = 5.25VEE300 pKa = 3.87ATEE303 pKa = 4.26LKK305 pKa = 10.69FEE307 pKa = 4.38ALQFRR312 pKa = 11.84YY313 pKa = 9.46HH314 pKa = 6.67SKK316 pKa = 9.31LTVEE320 pKa = 4.89CGLDD324 pKa = 3.45LAAQLYY330 pKa = 9.81EE331 pKa = 4.16LVEE334 pKa = 4.36CASDD338 pKa = 3.73FGVDD342 pKa = 3.37PTVAASVLQGLEE354 pKa = 3.96EE355 pKa = 3.77LAGAVEE361 pKa = 4.26EE362 pKa = 4.89TPCEE366 pKa = 3.84LVSWLNYY373 pKa = 10.19LGATIKK379 pKa = 10.6LLKK382 pKa = 10.19SYY384 pKa = 9.85EE385 pKa = 4.06RR386 pKa = 11.84EE387 pKa = 4.07FEE389 pKa = 5.79DD390 pKa = 3.6VAKK393 pKa = 10.2WDD395 pKa = 3.99DD396 pKa = 3.62VVDD399 pKa = 4.26CEE401 pKa = 4.7RR402 pKa = 11.84HH403 pKa = 5.72LEE405 pKa = 3.89PLQLHH410 pKa = 6.15CWEE413 pKa = 5.16IYY415 pKa = 10.2SPCC418 pKa = 4.06
Molecular weight: 45.18 kDa
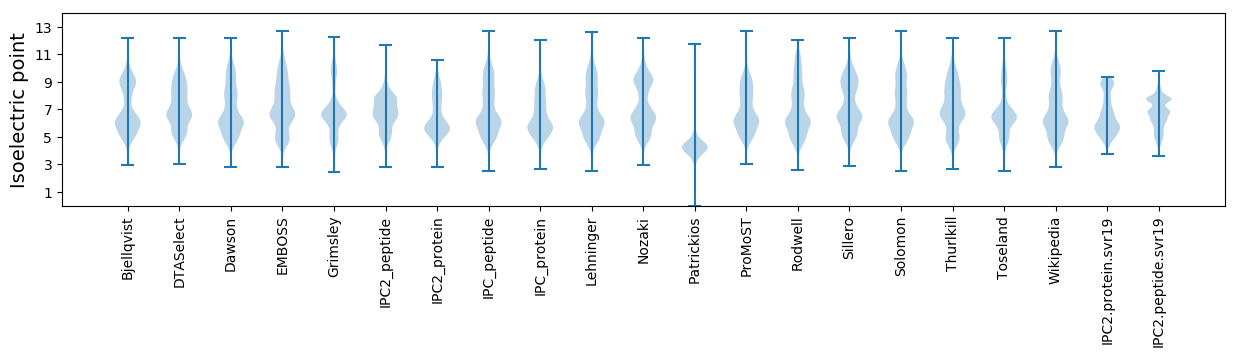
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V9YF75|A0A1V9YF75_9STRA Polycystin Cation Channel (PCC) Family OS=Achlya hypogyna OX=1202772 GN=ACHHYP_13415 PE=4 SV=1
MM1 pKa = 7.97PSPFPTSRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 9.43NVSAVEE17 pKa = 3.97ALCVLLNRR25 pKa = 11.84LAWPHH30 pKa = 6.44RR31 pKa = 11.84LGSMVSHH38 pKa = 7.45FGRR41 pKa = 11.84SRR43 pKa = 11.84EE44 pKa = 4.0ALSTIFNAALHH55 pKa = 6.38HH56 pKa = 5.92INEE59 pKa = 4.2RR60 pKa = 11.84FARR63 pKa = 11.84LLKK66 pKa = 10.06WDD68 pKa = 4.0DD69 pKa = 3.33RR70 pKa = 11.84RR71 pKa = 11.84LDD73 pKa = 3.83GRR75 pKa = 11.84WMAACAKK82 pKa = 10.01AIHH85 pKa = 6.64AKK87 pKa = 9.77GAPLDD92 pKa = 3.82SCIGFIDD99 pKa = 3.82GTVRR103 pKa = 11.84GICRR107 pKa = 11.84PKK109 pKa = 10.87NGVQRR114 pKa = 11.84AAYY117 pKa = 9.41NVYY120 pKa = 10.26KK121 pKa = 10.61VLQFQPGLTNN131 pKa = 3.62
MM1 pKa = 7.97PSPFPTSRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 9.43NVSAVEE17 pKa = 3.97ALCVLLNRR25 pKa = 11.84LAWPHH30 pKa = 6.44RR31 pKa = 11.84LGSMVSHH38 pKa = 7.45FGRR41 pKa = 11.84SRR43 pKa = 11.84EE44 pKa = 4.0ALSTIFNAALHH55 pKa = 6.38HH56 pKa = 5.92INEE59 pKa = 4.2RR60 pKa = 11.84FARR63 pKa = 11.84LLKK66 pKa = 10.06WDD68 pKa = 4.0DD69 pKa = 3.33RR70 pKa = 11.84RR71 pKa = 11.84LDD73 pKa = 3.83GRR75 pKa = 11.84WMAACAKK82 pKa = 10.01AIHH85 pKa = 6.64AKK87 pKa = 9.77GAPLDD92 pKa = 3.82SCIGFIDD99 pKa = 3.82GTVRR103 pKa = 11.84GICRR107 pKa = 11.84PKK109 pKa = 10.87NGVQRR114 pKa = 11.84AAYY117 pKa = 9.41NVYY120 pKa = 10.26KK121 pKa = 10.61VLQFQPGLTNN131 pKa = 3.62
Molecular weight: 14.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7479346 |
49 |
12321 |
519.4 |
57.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.161 ± 0.03 | 1.782 ± 0.01 |
5.538 ± 0.015 | 5.268 ± 0.022 |
3.749 ± 0.013 | 5.858 ± 0.029 |
2.766 ± 0.011 | 3.967 ± 0.014 |
4.353 ± 0.023 | 10.29 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.358 ± 0.008 | 2.988 ± 0.013 |
5.463 ± 0.022 | 3.768 ± 0.013 |
5.828 ± 0.023 | 6.777 ± 0.025 |
6.24 ± 0.026 | 7.73 ± 0.018 |
1.358 ± 0.008 | 2.757 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
